Guobao Wang
Patlak Parametric Image Estimation from Dynamic PET Using Diffusion Model Prior
Dec 22, 2025Abstract:Dynamic PET enables the quantitative estimation of physiology-related parameters and is widely utilized in research and increasingly adopted in clinical settings. Parametric imaging in dynamic PET requires kinetic modeling to estimate voxel-wise physiological parameters based on specific kinetic models. However, parametric images estimated through kinetic model fitting often suffer from low image quality due to the inherently ill-posed nature of the fitting process and the limited counts resulting from non-continuous data acquisition across multiple bed positions in whole-body PET. In this work, we proposed a diffusion model-based kinetic modeling framework for parametric image estimation, using the Patlak model as an example. The score function of the diffusion model was pre-trained on static total-body PET images and served as a prior for both Patlak slope and intercept images by leveraging their patch-wise similarity. During inference, the kinetic model was incorporated as a data-consistency constraint to guide the parametric image estimation. The proposed framework was evaluated on total-body dynamic PET datasets with different dose levels, demonstrating the feasibility and promising performance of the proposed framework in improving parametric image quality.
Systematic Review on Learning-based Spectral CT
Apr 15, 2023



Abstract:Spectral computed tomography (CT) has recently emerged as an advanced version of medical CT and significantly improves conventional (single-energy) CT. Spectral CT has two main forms: dual-energy computed tomography (DECT) and photon-counting computed tomography (PCCT), which offer image improvement, material decomposition, and feature quantification relative to conventional CT. However, the inherent challenges of spectral CT, evidenced by data and image artifacts, remain a bottleneck for clinical applications. To address these problems, machine learning techniques have been widely applied to spectral CT. In this review, we present the state-of-the-art data-driven techniques for spectral CT.
Neural KEM: A Kernel Method with Deep Coefficient Prior for PET Image Reconstruction
Jan 05, 2022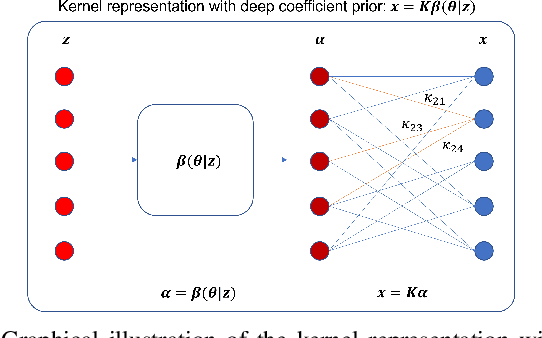
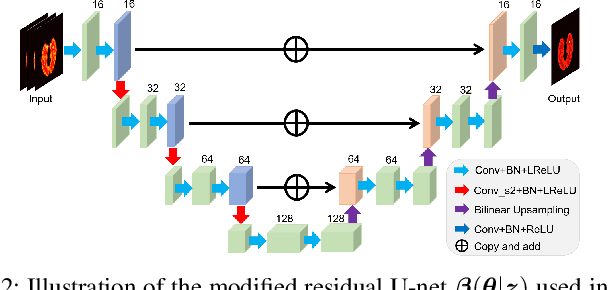
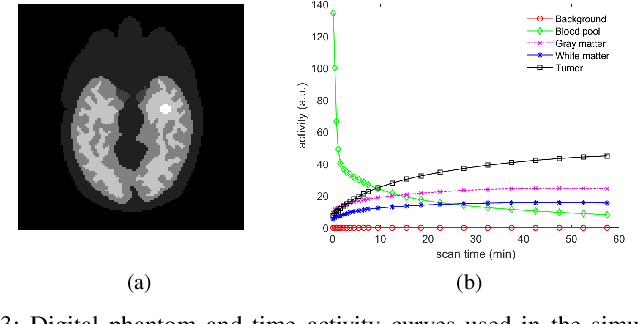
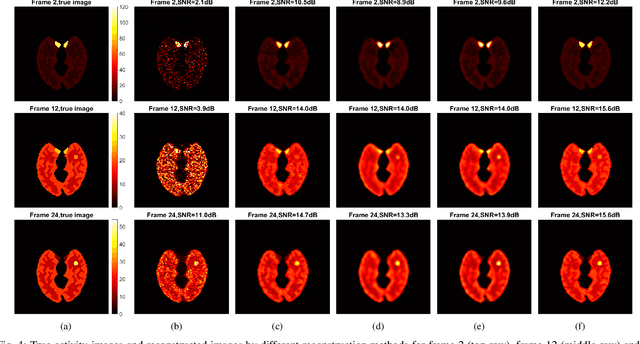
Abstract:Image reconstruction of low-count positron emission tomography (PET) data is challenging. Kernel methods address the challenge by incorporating image prior information in the forward model of iterative PET image reconstruction. The kernelized expectation-maximization (KEM) algorithm has been developed and demonstrated to be effective and easy to implement. A common approach for a further improvement of the kernel method would be adding an explicit regularization, which however leads to a complex optimization problem. In this paper, we propose an implicit regularization for the kernel method by using a deep coefficient prior, which represents the kernel coefficient image in the PET forward model using a convolutional neural-network. To solve the maximum-likelihood neural network-based reconstruction problem, we apply the principle of optimization transfer to derive a neural KEM algorithm. Each iteration of the algorithm consists of two separate steps: a KEM step for image update from the projection data and a deep-learning step in the image domain for updating the kernel coefficient image using the neural network. This optimization algorithm is guaranteed to monotonically increase the data likelihood. The results from computer simulations and real patient data have demonstrated that the neural KEM can outperform existing KEM and deep image prior methods.
Deep Kernel Representation for Image Reconstruction in PET
Oct 04, 2021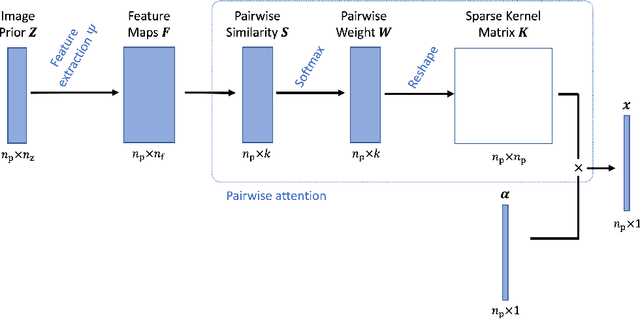
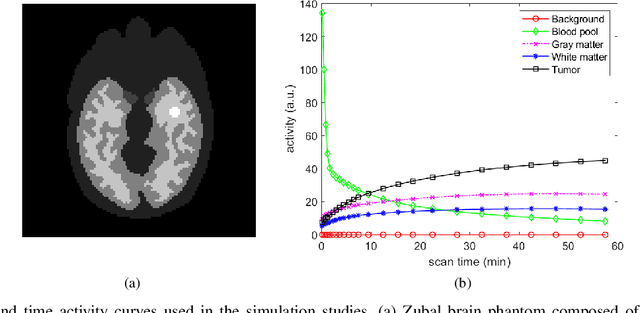
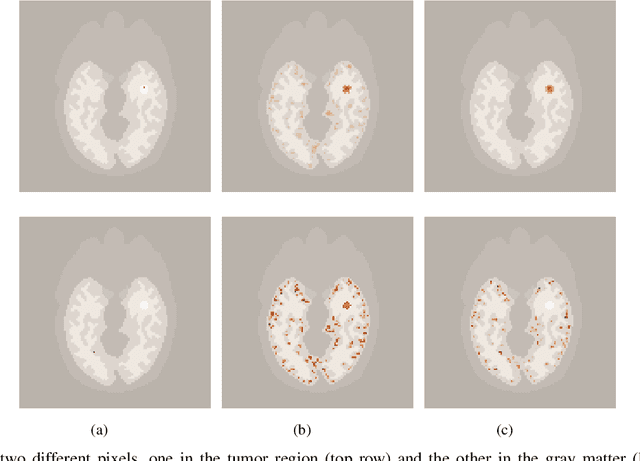
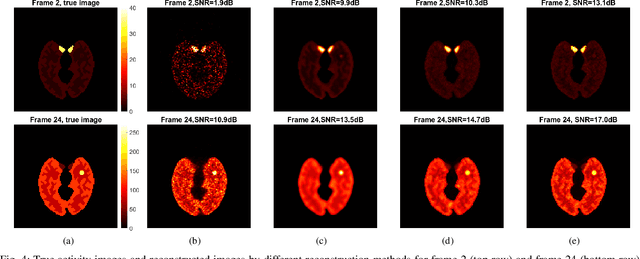
Abstract:Image reconstruction for positron emission tomography (PET) is challenging because of the ill-conditioned tomographic problem and low counting statistics. Kernel methods address this challenge by using kernel representation to incorporate image prior information in the forward model of iterative PET image reconstruction. Existing kernel methods construct the kernels commonly using an empirical process, which may lead to suboptimal performance. In this paper, we describe the equivalence between the kernel representation and a trainable neural network model. A deep kernel method is proposed by exploiting deep neural networks to enable an automated learning of an optimized kernel model. The proposed method is directly applicable to single subjects. The training process utilizes available image prior data to seek the best way to form a set of robust kernels optimally rather than empirically. The results from computer simulations and a real patient dataset demonstrate that the proposed deep kernel method can outperform existing kernel method and neural network method for dynamic PET image reconstruction.
Low-Dose CT Image Denoising Using Parallel-Clone Networks
May 14, 2020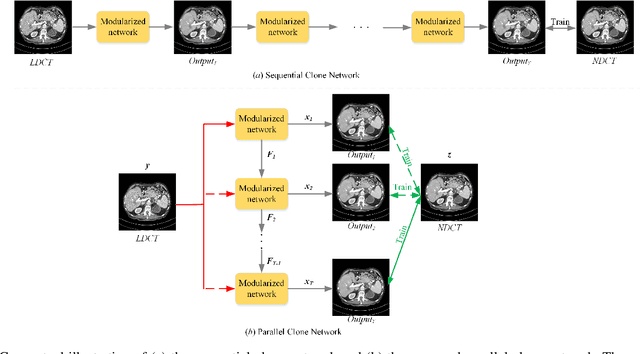
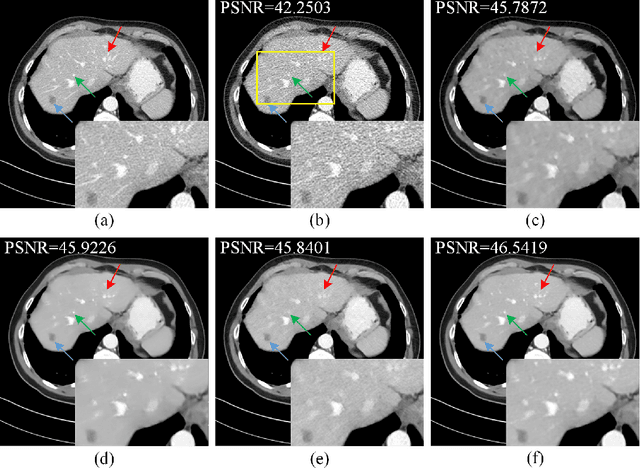
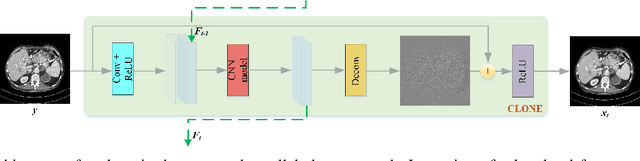
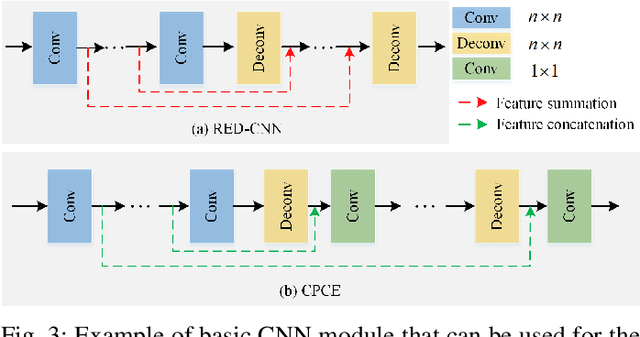
Abstract:Deep neural networks have a great potential to improve image denoising in low-dose computed tomography (LDCT). Popular ways to increase the network capacity include adding more layers or repeating a modularized clone model in a sequence. In such sequential architectures, the noisy input image and end output image are commonly used only once in the training model, which however limits the overall learning performance. In this paper, we propose a parallel-clone neural network method that utilizes a modularized network model and exploits the benefit of parallel input, parallel-output loss, and clone-toclone feature transfer. The proposed model keeps a similar or less number of unknown network weights as compared to conventional models but can accelerate the learning process significantly. The method was evaluated using the Mayo LDCT dataset and compared with existing deep learning models. The results show that the use of parallel input, parallel-output loss, and clone-to-clone feature transfer all can contribute to an accelerated convergence of deep learning and lead to improved image quality in testing. The parallel-clone network has been demonstrated promising for LDCT image denoising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge