Gonglei Shi
CreatiPoster: Towards Editable and Controllable Multi-Layer Graphic Design Generation
Jun 12, 2025Abstract:Graphic design plays a crucial role in both commercial and personal contexts, yet creating high-quality, editable, and aesthetically pleasing graphic compositions remains a time-consuming and skill-intensive task, especially for beginners. Current AI tools automate parts of the workflow, but struggle to accurately incorporate user-supplied assets, maintain editability, and achieve professional visual appeal. Commercial systems, like Canva Magic Design, rely on vast template libraries, which are impractical for replicate. In this paper, we introduce CreatiPoster, a framework that generates editable, multi-layer compositions from optional natural-language instructions or assets. A protocol model, an RGBA large multimodal model, first produces a JSON specification detailing every layer (text or asset) with precise layout, hierarchy, content and style, plus a concise background prompt. A conditional background model then synthesizes a coherent background conditioned on this rendered foreground layers. We construct a benchmark with automated metrics for graphic-design generation and show that CreatiPoster surpasses leading open-source approaches and proprietary commercial systems. To catalyze further research, we release a copyright-free corpus of 100,000 multi-layer designs. CreatiPoster supports diverse applications such as canvas editing, text overlay, responsive resizing, multilingual adaptation, and animated posters, advancing the democratization of AI-assisted graphic design. Project homepage: https://github.com/graphic-design-ai/creatiposter
Marginal loss and exclusion loss for partially supervised multi-organ segmentation
Jul 08, 2020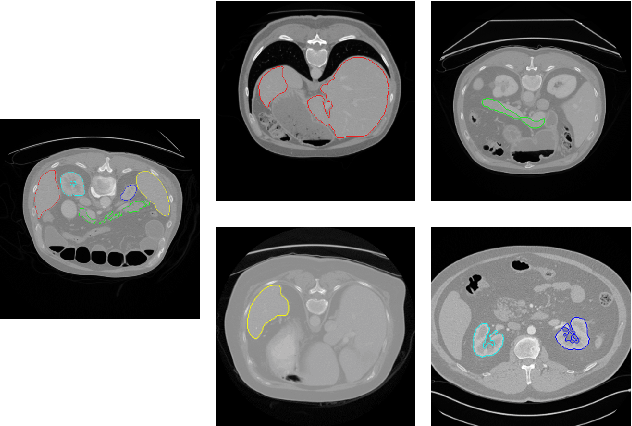
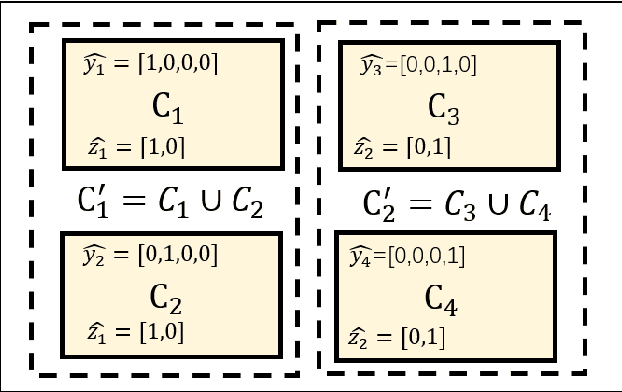
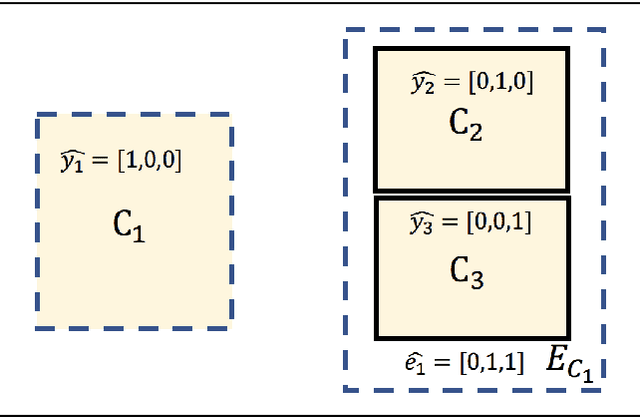
Abstract:Annotating multiple organs in medical images is both costly and time-consuming; therefore, existing multi-organ datasets with labels are often low in sample size and mostly partially labeled, that is, a dataset has a few organs labeled but not all organs. In this paper, we investigate how to learn a single multi-organ segmentation network from a union of such datasets. To this end, we propose two types of novel loss function, particularly designed for this scenario: (i) marginal loss and (ii) exclusion loss. Because the background label for a partially labeled image is, in fact, a `merged' label of all unlabelled organs and `true' background (in the sense of full labels), the probability of this `merged' background label is a marginal probability, summing the relevant probabilities before merging. This marginal probability can be plugged into any existing loss function (such as cross entropy loss, Dice loss, etc.) to form a marginal loss. Leveraging the fact that the organs are non-overlapping, we propose the exclusion loss to gauge the dissimilarity between labeled organs and the estimated segmentation of unlabelled organs. Experiments on a union of five benchmark datasets in multi-organ segmentation of liver, spleen, left and right kidneys, and pancreas demonstrate that using our newly proposed loss functions brings a conspicuous performance improvement for state-of-the-art methods without introducing any extra computation.
Encoding CT Anatomy Knowledge for Unpaired Chest X-ray Image Decomposition
Sep 16, 2019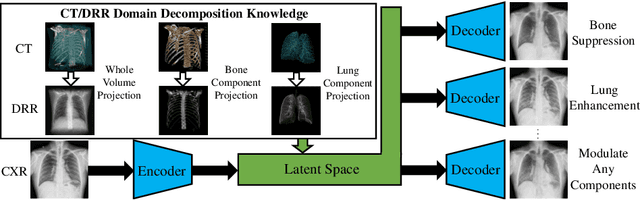
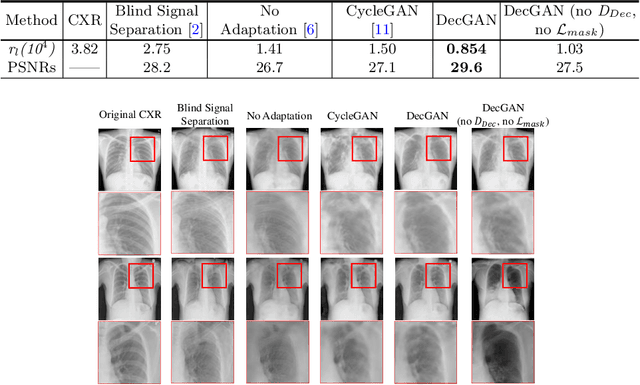
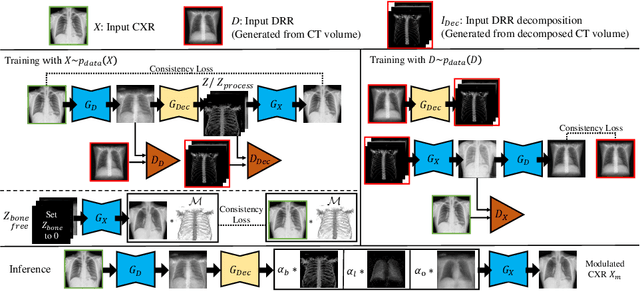
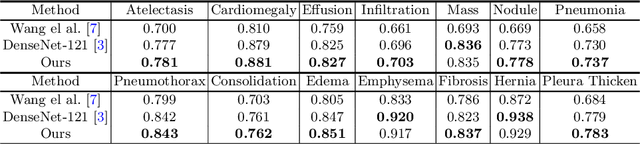
Abstract:Although chest X-ray (CXR) offers a 2D projection with overlapped anatomies, it is widely used for clinical diagnosis. There is clinical evidence supporting that decomposing an X-ray image into different components (e.g., bone, lung and soft tissue) improves diagnostic value. We hereby propose a decomposition generative adversarial network (DecGAN) to anatomically decompose a CXR image but with unpaired data. We leverage the anatomy knowledge embedded in CT, which features a 3D volume with clearly visible anatomies. Our key idea is to embed CT priori decomposition knowledge into the latent space of unpaired CXR autoencoder. Specifically, we train DecGAN with a decomposition loss, adversarial losses, cycle-consistency losses and a mask loss to guarantee that the decomposed results of the latent space preserve realistic body structures. Extensive experiments demonstrate that DecGAN provides superior unsupervised CXR bone suppression results and the feasibility of modulating CXR components by latent space disentanglement. Furthermore, we illustrate the diagnostic value of DecGAN and demonstrate that it outperforms the state-of-the-art approaches in terms of predicting 11 out of 14 common lung diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge