Geoffrey Fox
Leveraging Exogenous Signals for Hydrology Time Series Forecasting
Nov 14, 2025Abstract:Recent advances in time series research facilitate the development of foundation models. While many state-of-the-art time series foundation models have been introduced, few studies examine their effectiveness in specific downstream applications in physical science. This work investigates the role of integrating domain knowledge into time series models for hydrological rainfall-runoff modeling. Using the CAMELS-US dataset, which includes rainfall and runoff data from 671 locations with six time series streams and 30 static features, we compare baseline and foundation models. Results demonstrate that models incorporating comprehensive known exogenous inputs outperform more limited approaches, including foundation models. Notably, incorporating natural annual periodic time series contribute the most significant improvements.
An MLCommons Scientific Benchmarks Ontology
Nov 06, 2025



Abstract:Scientific machine learning research spans diverse domains and data modalities, yet existing benchmark efforts remain siloed and lack standardization. This makes novel and transformative applications of machine learning to critical scientific use-cases more fragmented and less clear in pathways to impact. This paper introduces an ontology for scientific benchmarking developed through a unified, community-driven effort that extends the MLCommons ecosystem to cover physics, chemistry, materials science, biology, climate science, and more. Building on prior initiatives such as XAI-BENCH, FastML Science Benchmarks, PDEBench, and the SciMLBench framework, our effort consolidates a large set of disparate benchmarks and frameworks into a single taxonomy of scientific, application, and system-level benchmarks. New benchmarks can be added through an open submission workflow coordinated by the MLCommons Science Working Group and evaluated against a six-category rating rubric that promotes and identifies high-quality benchmarks, enabling stakeholders to select benchmarks that meet their specific needs. The architecture is extensible, supporting future scientific and AI/ML motifs, and we discuss methods for identifying emerging computing patterns for unique scientific workloads. The MLCommons Science Benchmarks Ontology provides a standardized, scalable foundation for reproducible, cross-domain benchmarking in scientific machine learning. A companion webpage for this work has also been developed as the effort evolves: https://mlcommons-science.github.io/benchmark/
IrrMap: A Large-Scale Comprehensive Dataset for Irrigation Method Mapping
May 13, 2025



Abstract:We introduce IrrMap, the first large-scale dataset (1.1 million patches) for irrigation method mapping across regions. IrrMap consists of multi-resolution satellite imagery from LandSat and Sentinel, along with key auxiliary data such as crop type, land use, and vegetation indices. The dataset spans 1,687,899 farms and 14,117,330 acres across multiple western U.S. states from 2013 to 2023, providing a rich and diverse foundation for irrigation analysis and ensuring geospatial alignment and quality control. The dataset is ML-ready, with standardized 224x224 GeoTIFF patches, the multiple input modalities, carefully chosen train-test-split data, and accompanying dataloaders for seamless deep learning model training andbenchmarking in irrigation mapping. The dataset is also accompanied by a complete pipeline for dataset generation, enabling researchers to extend IrrMap to new regions for irrigation data collection or adapt it with minimal effort for other similar applications in agricultural and geospatial analysis. We also analyze the irrigation method distribution across crop groups, spatial irrigation patterns (using Shannon diversity indices), and irrigated area variations for both LandSat and Sentinel, providing insights into regional and resolution-based differences. To promote further exploration, we openly release IrrMap, along with the derived datasets, benchmark models, and pipeline code, through a GitHub repository: https://github.com/Nibir088/IrrMap and Data repository: https://huggingface.co/Nibir/IrrMap, providing comprehensive documentation and implementation details.
Surrogate modeling of Cellular-Potts Agent-Based Models as a segmentation task using the U-Net neural network architecture
May 05, 2025Abstract:The Cellular-Potts model is a powerful and ubiquitous framework for developing computational models for simulating complex multicellular biological systems. Cellular-Potts models (CPMs) are often computationally expensive due to the explicit modeling of interactions among large numbers of individual model agents and diffusive fields described by partial differential equations (PDEs). In this work, we develop a convolutional neural network (CNN) surrogate model using a U-Net architecture that accounts for periodic boundary conditions. We use this model to accelerate the evaluation of a mechanistic CPM previously used to investigate \textit{in vitro} vasculogenesis. The surrogate model was trained to predict 100 computational steps ahead (Monte-Carlo steps, MCS), accelerating simulation evaluations by a factor of 590 times compared to CPM code execution. Over multiple recursive evaluations, our model effectively captures the emergent behaviors demonstrated by the original Cellular-Potts model of such as vessel sprouting, extension and anastomosis, and contraction of vascular lacunae. This approach demonstrates the potential for deep learning to serve as efficient surrogate models for CPM simulations, enabling faster evaluation of computationally expensive CPM of biological processes at greater spatial and temporal scales.
Building Machine Learning Challenges for Anomaly Detection in Science
Mar 03, 2025
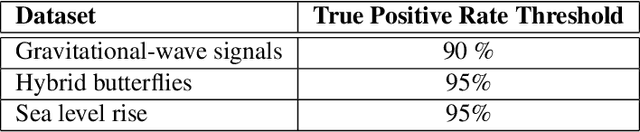


Abstract:Scientific discoveries are often made by finding a pattern or object that was not predicted by the known rules of science. Oftentimes, these anomalous events or objects that do not conform to the norms are an indication that the rules of science governing the data are incomplete, and something new needs to be present to explain these unexpected outliers. The challenge of finding anomalies can be confounding since it requires codifying a complete knowledge of the known scientific behaviors and then projecting these known behaviors on the data to look for deviations. When utilizing machine learning, this presents a particular challenge since we require that the model not only understands scientific data perfectly but also recognizes when the data is inconsistent and out of the scope of its trained behavior. In this paper, we present three datasets aimed at developing machine learning-based anomaly detection for disparate scientific domains covering astrophysics, genomics, and polar science. We present the different datasets along with a scheme to make machine learning challenges around the three datasets findable, accessible, interoperable, and reusable (FAIR). Furthermore, we present an approach that generalizes to future machine learning challenges, enabling the possibility of large, more compute-intensive challenges that can ultimately lead to scientific discovery.
Scalable Cosmic AI Inference using Cloud Serverless Computing with FMI
Jan 08, 2025



Abstract:Large-scale astronomical image data processing and prediction is essential for astronomers, providing crucial insights into celestial objects, the universe's history, and its evolution. While modern deep learning models offer high predictive accuracy, they often demand substantial computational resources, making them resource-intensive and limiting accessibility. We introduce the Cloud-based Astronomy Inference (CAI) framework to address these challenges. This scalable solution integrates pre-trained foundation models with serverless cloud infrastructure through a Function-as-a-Service (FaaS) Message Interface (FMI). CAI enables efficient and scalable inference on astronomical images without extensive hardware. Using a foundation model for redshift prediction as a case study, our extensive experiments cover user devices, HPC (High-Performance Computing) servers, and Cloud. CAI's significant scalability improvement on large data sizes provides an accessible and effective tool for the astronomy community. The code is accessible at https://github.com/UVA-MLSys/AI-for-Astronomy.
Zephyr quantum-assisted hierarchical Calo4pQVAE for particle-calorimeter interactions
Dec 06, 2024Abstract:With the approach of the High Luminosity Large Hadron Collider (HL-LHC) era set to begin particle collisions by the end of this decade, it is evident that the computational demands of traditional collision simulation methods are becoming increasingly unsustainable. Existing approaches, which rely heavily on first-principles Monte Carlo simulations for modeling event showers in calorimeters, are projected to require millions of CPU-years annually -- far exceeding current computational capacities. This bottleneck presents an exciting opportunity for advancements in computational physics by integrating deep generative models with quantum simulations. We propose a quantum-assisted hierarchical deep generative surrogate founded on a variational autoencoder (VAE) in combination with an energy conditioned restricted Boltzmann machine (RBM) embedded in the model's latent space as a prior. By mapping the topology of D-Wave's Zephyr quantum annealer (QA) into the nodes and couplings of a 4-partite RBM, we leverage quantum simulation to accelerate our shower generation times significantly. To evaluate our framework, we use Dataset 2 of the CaloChallenge 2022. Through the integration of classical computation and quantum simulation, this hybrid framework paves way for utilizing large-scale quantum simulations as priors in deep generative models.
Science Time Series: Deep Learning in Hydrology
Oct 19, 2024Abstract:This research is part of a systematic study of scientific time series. In the last three years, hundreds of papers and over fifty new deep-learning models have been described for time series models. These mainly focus on the key aspect of time dependence, whereas in some scientific time series, the situation is more complex with multiple locations, each location having multiple observed and target time-dependent streams and multiple exogenous (known) properties that are either constant or time-dependent. Here, we analyze the hydrology time series using the CAMELS and Caravan global datasets on catchment rainfall and runoff. Together, these have up to 6 observed streams and up to 209 static parameters defined at each of about 8000 locations. This analysis is fully open source with a Jupyter Notebook running on Google Colab for both an LSTM-based analysis and the data engineering preprocessing. Our goal is to investigate the importance of exogenous data, which we look at using eight different choices on representative hydrology tasks. Increasing the exogenous information significantly improves the data representation, with the mean square error decreasing to 60% of its initial value in the largest dataset examined. We present the initial results of studies of other deep-learning neural network architectures where the approaches that can use the full observed and exogenous observations outperform less flexible methods, including Foundation models. Using the natural annual periodic exogenous time series produces the largest impact, but the static and other periodic exogenous streams are also important. Our analysis is intended to be valuable as an educational resource and benchmark.
Study of Dropout in PointPillars with 3D Object Detection
Sep 01, 2024Abstract:3D object detection is critical for autonomous driving, leveraging deep learning techniques to interpret LiDAR data. The PointPillars architecture is a prominent model in this field, distinguished by its efficient use of LiDAR data. This study provides an analysis of enhancing the performance of PointPillars model under various dropout rates to address overfitting and improve model generalization. Dropout, a regularization technique, involves randomly omitting neurons during training, compelling the network to learn robust and diverse features. We systematically compare the effects of different enhancement techniques on the model's regression performance during training and its accuracy, measured by Average Precision (AP) and Average Orientation Similarity (AOS). Our findings offer insights into the optimal enhancements, contributing to improved 3D object detection in autonomous driving applications.
Time Series Foundation Models and Deep Learning Architectures for Earthquake Temporal and Spatial Nowcasting
Aug 21, 2024



Abstract:Advancing the capabilities of earthquake nowcasting, the real-time forecasting of seismic activities remains a crucial and enduring objective aimed at reducing casualties. This multifaceted challenge has recently gained attention within the deep learning domain, facilitated by the availability of extensive, long-term earthquake datasets. Despite significant advancements, existing literature on earthquake nowcasting lacks comprehensive evaluations of pre-trained foundation models and modern deep learning architectures. These architectures, such as transformers or graph neural networks, uniquely focus on different aspects of data, including spatial relationships, temporal patterns, and multi-scale dependencies. This paper addresses the mentioned gap by analyzing different architectures and introducing two innovation approaches called MultiFoundationQuake and GNNCoder. We formulate earthquake nowcasting as a time series forecasting problem for the next 14 days within 0.1-degree spatial bins in Southern California, spanning from 1986 to 2024. Earthquake time series is forecasted as a function of logarithm energy released by quakes. Our comprehensive evaluation employs several key performance metrics, notably Nash-Sutcliffe Efficiency and Mean Squared Error, over time in each spatial region. The results demonstrate that our introduced models outperform other custom architectures by effectively capturing temporal-spatial relationships inherent in seismic data. The performance of existing foundation models varies significantly based on the pre-training datasets, emphasizing the need for careful dataset selection. However, we introduce a new general approach termed MultiFoundationPattern that combines a bespoke pattern with foundation model results handled as auxiliary streams. In the earthquake case, the resultant MultiFoundationQuake model achieves the best overall performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge