Fei Liao
RoboMIND 2.0: A Multimodal, Bimanual Mobile Manipulation Dataset for Generalizable Embodied Intelligence
Dec 31, 2025Abstract:While data-driven imitation learning has revolutionized robotic manipulation, current approaches remain constrained by the scarcity of large-scale, diverse real-world demonstrations. Consequently, the ability of existing models to generalize across long-horizon bimanual tasks and mobile manipulation in unstructured environments remains limited. To bridge this gap, we present RoboMIND 2.0, a comprehensive real-world dataset comprising over 310K dual-arm manipulation trajectories collected across six distinct robot embodiments and 739 complex tasks. Crucially, to support research in contact-rich and spatially extended tasks, the dataset incorporates 12K tactile-enhanced episodes and 20K mobile manipulation trajectories. Complementing this physical data, we construct high-fidelity digital twins of our real-world environments, releasing an additional 20K-trajectory simulated dataset to facilitate robust sim-to-real transfer. To fully exploit the potential of RoboMIND 2.0, we propose MIND-2 system, a hierarchical dual-system frame-work optimized via offline reinforcement learning. MIND-2 integrates a high-level semantic planner (MIND-2-VLM) to decompose abstract natural language instructions into grounded subgoals, coupled with a low-level Vision-Language-Action executor (MIND-2-VLA), which generates precise, proprioception-aware motor actions.
Resolving Knowledge Conflicts in Domain-specific Data Selection: A Case Study on Medical Instruction-tuning
May 28, 2025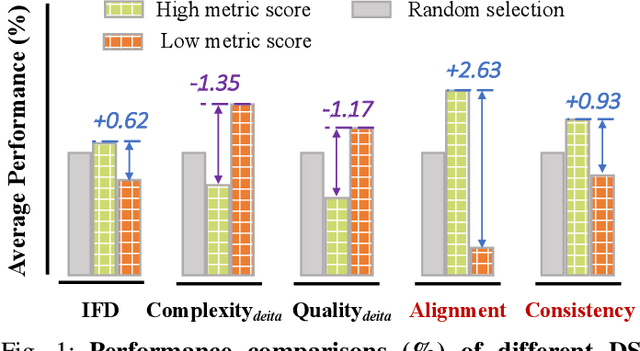
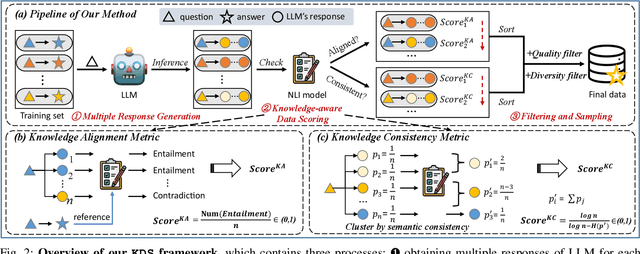
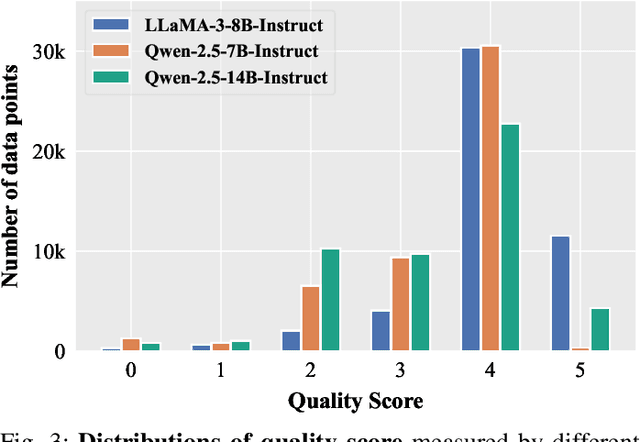
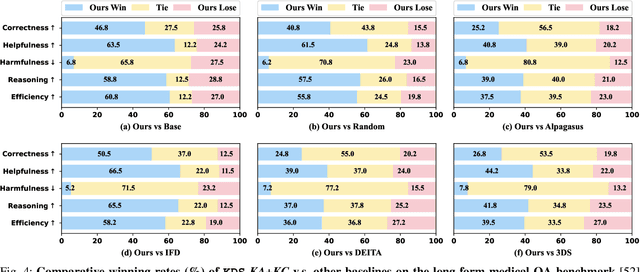
Abstract:Domain-specific instruction-tuning has become the defacto standard for improving the performance of large language models (LLMs) in specialized applications, e.g., medical question answering. Since the instruction-tuning dataset might contain redundant or low-quality data, data selection (DS) is usually required to maximize the data efficiency. Despite the successes in the general domain, current DS methods often struggle to select the desired data for domain-specific instruction-tuning. One of the main reasons is that they neglect the impact of knowledge conflicts, i.e., the discrepancy between LLMs' pretrained knowledge and context knowledge of instruction data, which could damage LLMs' prior abilities and lead to hallucination. To this end, we propose a simple-yet-effective Knowledge-aware Data Selection (namely KDS) framework to select the domain-specific instruction-tuning data that meets LLMs' actual needs. The core of KDS is to leverage two knowledge-aware metrics for quantitatively measuring knowledge conflicts from two aspects: context-memory knowledge alignment and intra-memory knowledge consistency. By filtering the data with large knowledge conflicts and sampling the high-quality and diverse data, KDS can effectively stimulate the LLMs' abilities and achieve better domain-specific performance. Taking the medical domain as the testbed, we conduct extensive experiments and empirically prove that KDS surpasses the other baselines and brings significant and consistent performance gains among all LLMs. More encouragingly, KDS effectively improves the model generalization and alleviates the hallucination problem.
Selective Information Passing for MR/CT Image Segmentation
Oct 10, 2020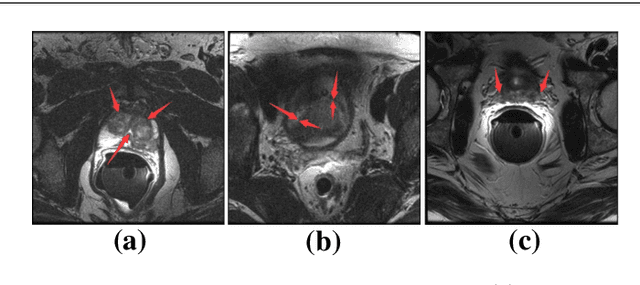
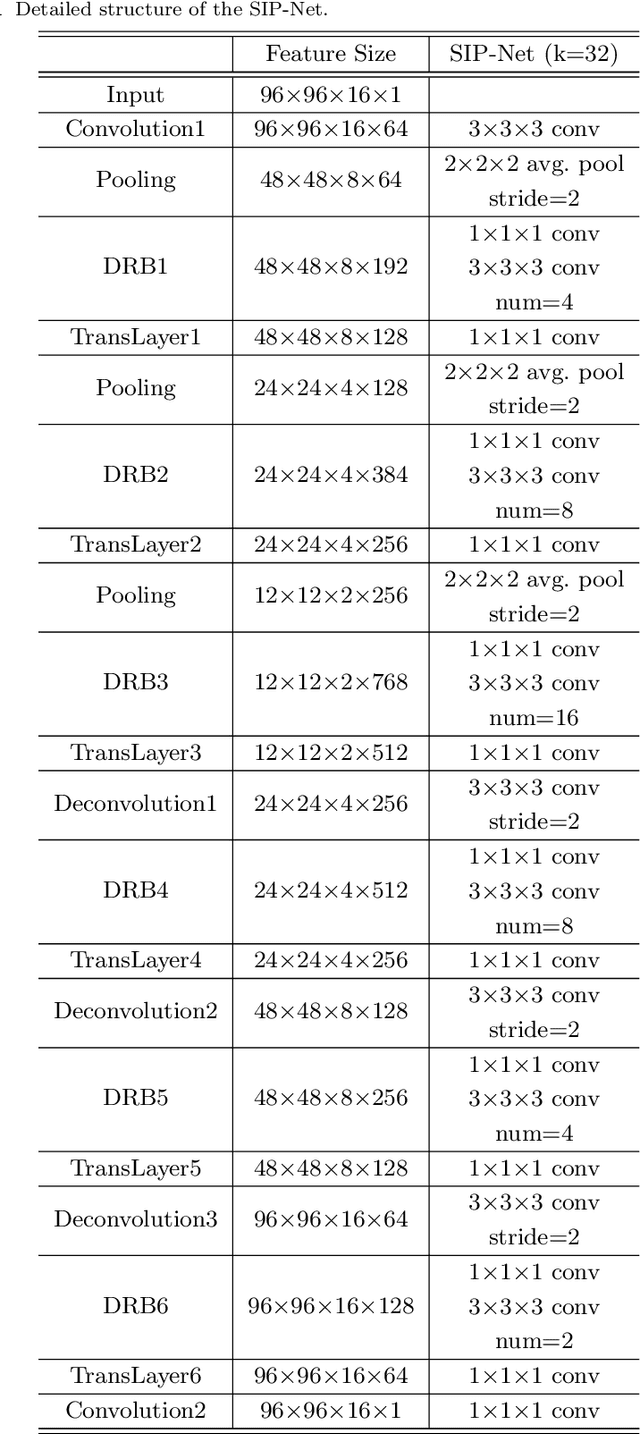
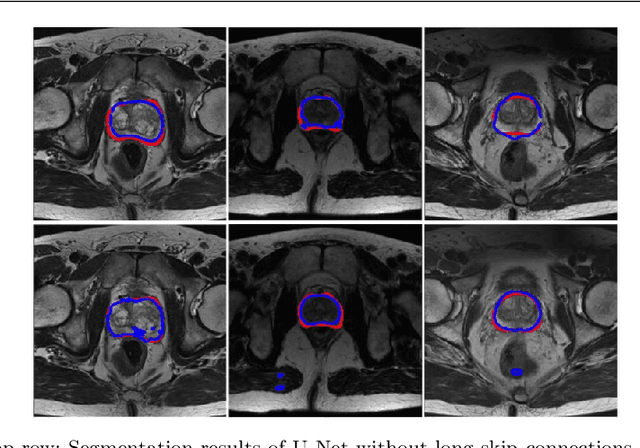
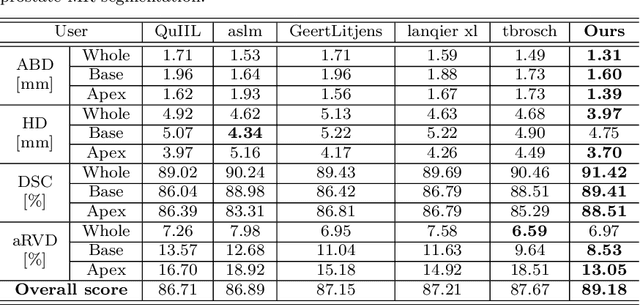
Abstract:Automated medical image segmentation plays an important role in many clinical applications, which however is a very challenging task, due to complex background texture, lack of clear boundary and significant shape and texture variation between images. Many researchers proposed an encoder-decoder architecture with skip connections to combine low-level feature maps from the encoder path with high-level feature maps from the decoder path for automatically segmenting medical images. The skip connections have been shown to be effective in recovering fine-grained details of the target objects and may facilitate the gradient back-propagation. However, not all the feature maps transmitted by those connections contribute positively to the network performance. In this paper, to adaptively select useful information to pass through those skip connections, we propose a novel 3D network with self-supervised function, named selective information passing network (SIP-Net). We evaluate our proposed model on the MICCAI Prostate MR Image Segmentation 2012 Grant Challenge dataset, TCIA Pancreas CT-82 and MICCAI 2017 Liver Tumor Segmentation (LiTS) Challenge dataset. The experimental results across these data sets show that our model achieved improved segmentation results and outperformed other state-of-the-art methods. The source code of this work is available at https://github.com/ahukui/SIPNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge