David Zimmerer
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
CRADL: Contrastive Representations for Unsupervised Anomaly Detection and Localization
Jan 05, 2023



Abstract:Unsupervised anomaly detection in medical imaging aims to detect and localize arbitrary anomalies without requiring annotated anomalous data during training. Often, this is achieved by learning a data distribution of normal samples and detecting anomalies as regions in the image which deviate from this distribution. Most current state-of-the-art methods use latent variable generative models operating directly on the images. However, generative models have been shown to mostly capture low-level features, s.a. pixel-intensities, instead of rich semantic features, which also applies to their representations. We circumvent this problem by proposing CRADL whose core idea is to model the distribution of normal samples directly in the low-dimensional representation space of an encoder trained with a contrastive pretext-task. By utilizing the representations of contrastive learning, we aim to fix the over-fixation on low-level features and learn more semantic-rich representations. Our experiments on anomaly detection and localization tasks using three distinct evaluation datasets show that 1) contrastive representations are superior to representations of generative latent variable models and 2) the CRADL framework shows competitive or superior performance to state-of-the-art.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Continuous-Time Deep Glioma Growth Models
Jul 02, 2021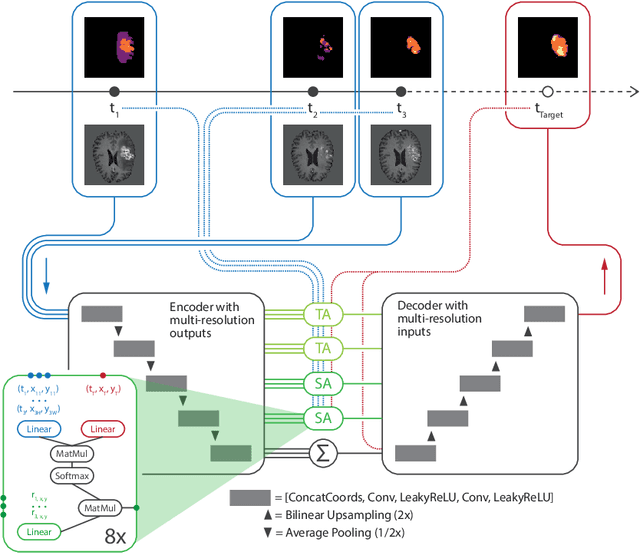
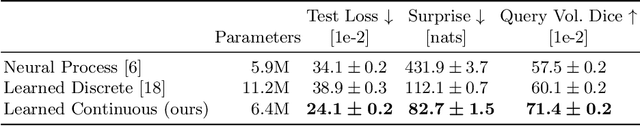

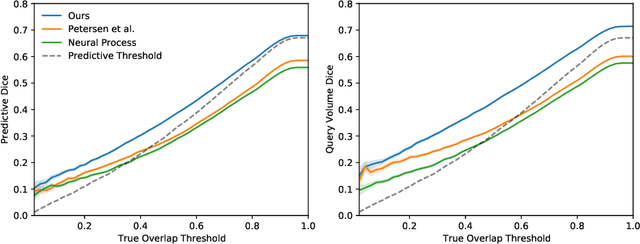
Abstract:The ability to estimate how a tumor might evolve in the future could have tremendous clinical benefits, from improved treatment decisions to better dose distribution in radiation therapy. Recent work has approached the glioma growth modeling problem via deep learning and variational inference, thus learning growth dynamics entirely from a real patient data distribution. So far, this approach was constrained to predefined image acquisition intervals and sequences of fixed length, which limits its applicability in more realistic scenarios. We overcome these limitations by extending Neural Processes, a class of conditional generative models for stochastic time series, with a hierarchical multi-scale representation encoding including a spatio-temporal attention mechanism. The result is a learned growth model that can be conditioned on an arbitrary number of observations, and that can produce a distribution of temporally consistent growth trajectories on a continuous time axis. On a dataset of 379 patients, the approach successfully captures both global and finer-grained variations in the images, exhibiting superior performance compared to other learned growth models.
GP-ConvCNP: Better Generalization for Convolutional Conditional Neural Processes on Time Series Data
Jun 11, 2021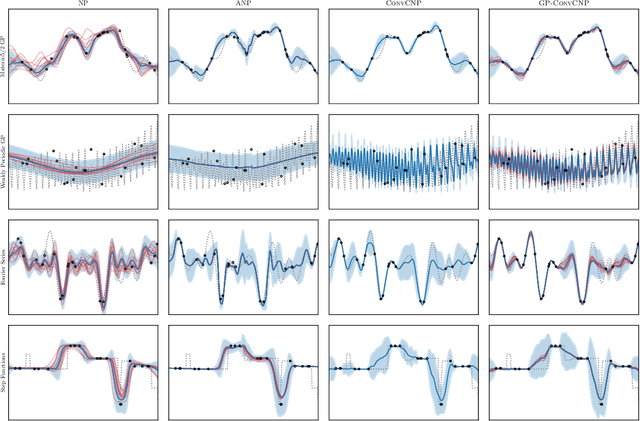
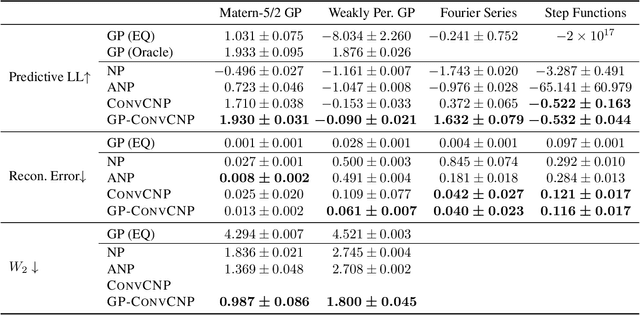
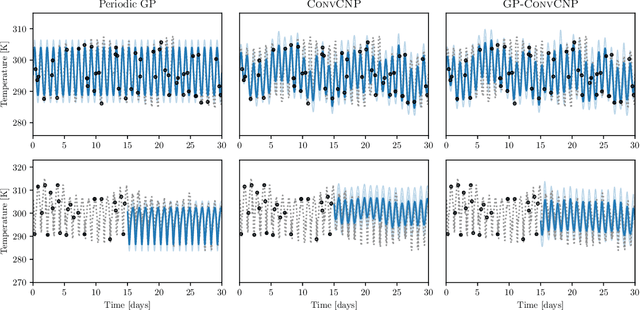
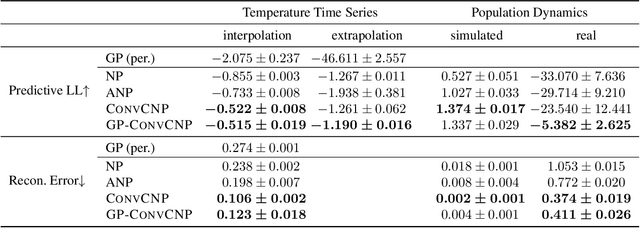
Abstract:Neural Processes (NPs) are a family of conditional generative models that are able to model a distribution over functions, in a way that allows them to perform predictions at test time conditioned on a number of context points. A recent addition to this family, Convolutional Conditional Neural Processes (ConvCNP), have shown remarkable improvement in performance over prior art, but we find that they sometimes struggle to generalize when applied to time series data. In particular, they are not robust to distribution shifts and fail to extrapolate observed patterns into the future. By incorporating a Gaussian Process into the model, we are able to remedy this and at the same time improve performance within distribution. As an added benefit, the Gaussian Process reintroduces the possibility to sample from the model, a key feature of other members in the NP family.
The Federated Tumor Segmentation (FeTS) Challenge
May 14, 2021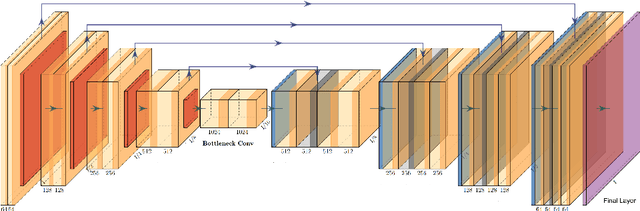
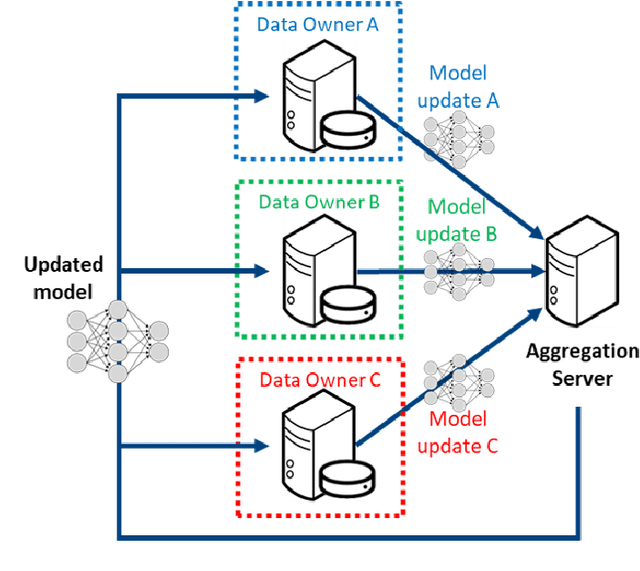
Abstract:This manuscript describes the first challenge on Federated Learning, namely the Federated Tumor Segmentation (FeTS) challenge 2021. International challenges have become the standard for validation of biomedical image analysis methods. However, the actual performance of participating (even the winning) algorithms on "real-world" clinical data often remains unclear, as the data included in challenges are usually acquired in very controlled settings at few institutions. The seemingly obvious solution of just collecting increasingly more data from more institutions in such challenges does not scale well due to privacy and ownership hurdles. Towards alleviating these concerns, we are proposing the FeTS challenge 2021 to cater towards both the development and the evaluation of models for the segmentation of intrinsically heterogeneous (in appearance, shape, and histology) brain tumors, namely gliomas. Specifically, the FeTS 2021 challenge uses clinically acquired, multi-institutional magnetic resonance imaging (MRI) scans from the BraTS 2020 challenge, as well as from various remote independent institutions included in the collaborative network of a real-world federation (https://www.fets.ai/). The goals of the FeTS challenge are directly represented by the two included tasks: 1) the identification of the optimal weight aggregation approach towards the training of a consensus model that has gained knowledge via federated learning from multiple geographically distinct institutions, while their data are always retained within each institution, and 2) the federated evaluation of the generalizability of brain tumor segmentation models "in the wild", i.e. on data from institutional distributions that were not part of the training datasets.
A Case for the Score: Identifying Image Anomalies using Variational Autoencoder Gradients
Nov 28, 2019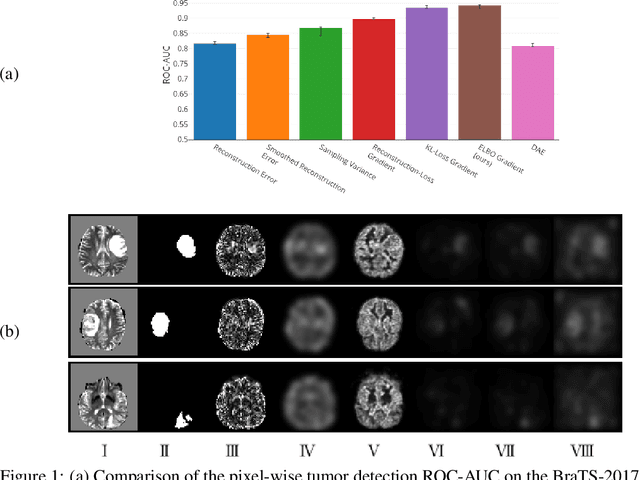
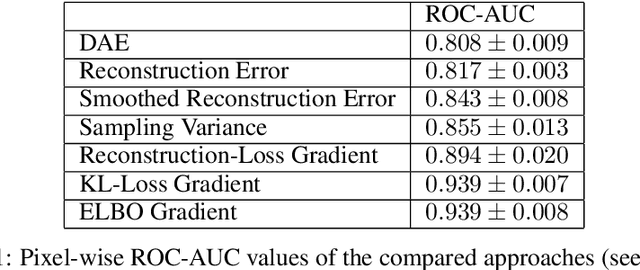
Abstract:Through training on unlabeled data, anomaly detection has the potential to impact computer-aided diagnosis by outlining suspicious regions. Previous work on deep-learning-based anomaly detection has primarily focused on the reconstruction error. We argue instead, that pixel-wise anomaly ratings derived from a Variational Autoencoder based score approximation yield a theoretically better grounded and more faithful estimate. In our experiments, Variational Autoencoder gradient-based rating outperforms other approaches on unsupervised pixel-wise tumor detection on the BraTS-2017 dataset with a ROC-AUC of 0.94.
High- and Low-level image component decomposition using VAEs for improved reconstruction and anomaly detection
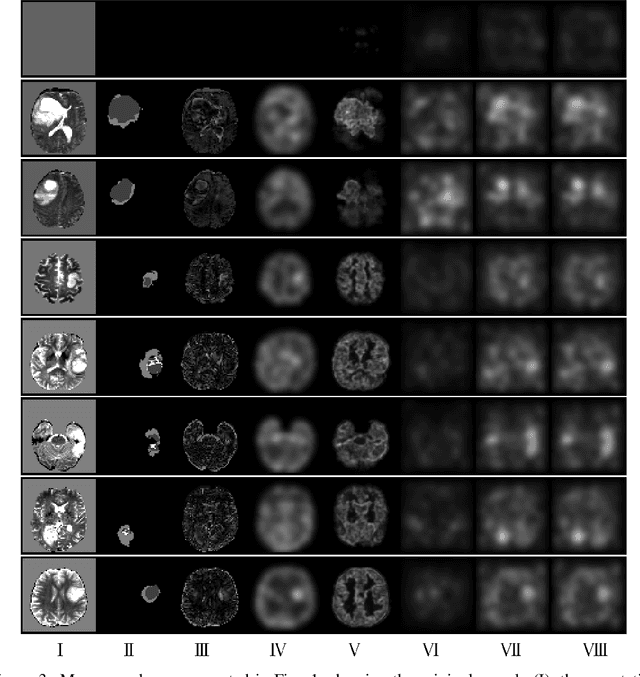
Nov 27, 2019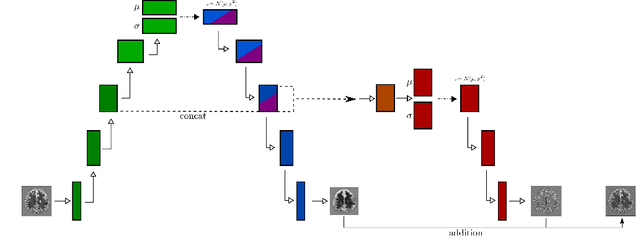
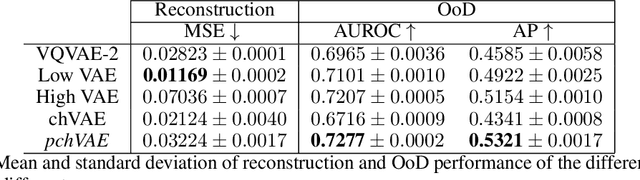
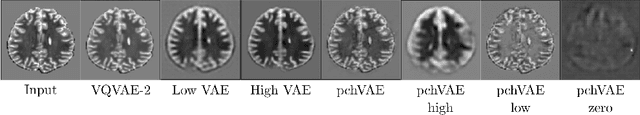
Abstract:Variational Auto-Encoders have often been used for unsupervised pretraining, feature extraction and out-of-distribution and anomaly detection in the medical field. However, VAEs often lack the ability to produce sharp images and learn high-level features. We propose to alleviate these issues by adding a new branch to conditional hierarchical VAEs. This enforces a division between higher-level and lower-level features. Despite the additional computational overhead compared to a normal VAE it results in sharper and better reconstructions and can capture the data distribution similarly well (indicated by a similar or slightly better OoD detection performance).
Unsupervised Anomaly Localization using Variational Auto-Encoders
Jul 11, 2019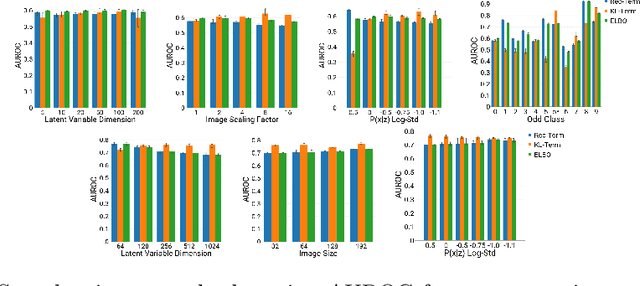
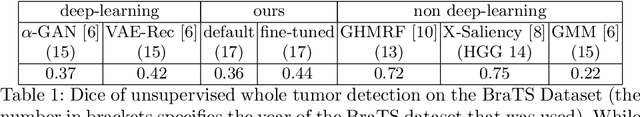
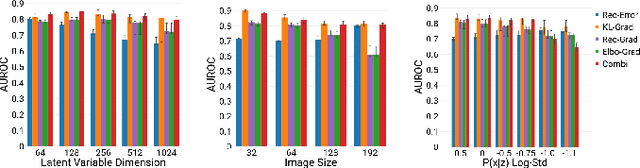
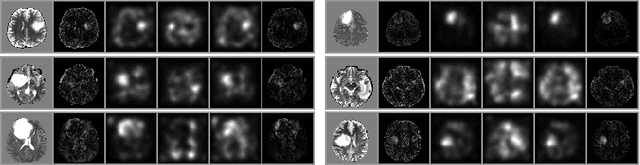
Abstract:An assumption-free automatic check of medical images for potentially overseen anomalies would be a valuable assistance for a radiologist. Deep learning and especially Variational Auto-Encoders (VAEs) have shown great potential in the unsupervised learning of data distributions. In principle, this allows for such a check and even the localization of parts in the image that are most suspicious. Currently, however, the reconstruction-based localization by design requires adjusting the model architecture to the specific problem looked at during evaluation. This contradicts the principle of building assumption-free models. We propose complementing the localization part with a term derived from the Kullback-Leibler (KL)-divergence. For validation, we perform a series of experiments on FashionMNIST as well as on a medical task including >1000 healthy and >250 brain tumor patients. Results show that the proposed formalism outperforms the state of the art VAE-based localization of anomalies across many hyperparameter settings and also shows a competitive max performance.
Context-encoding Variational Autoencoder for Unsupervised Anomaly Detection
Dec 14, 2018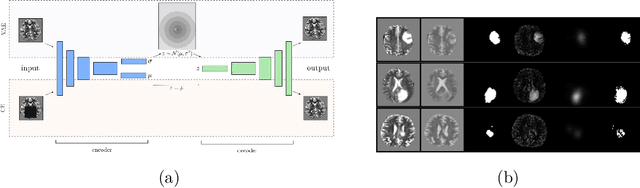
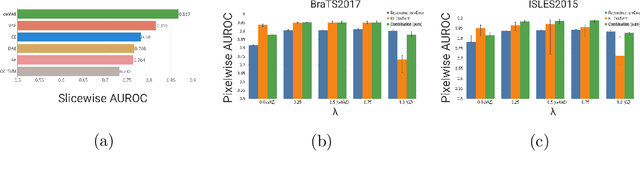
Abstract:Unsupervised learning can leverage large-scale data sources without the need for annotations. In this context, deep learning-based auto encoders have shown great potential in detecting anomalies in medical images. However, state-of-the-art anomaly scores are still based on the reconstruction error, which lacks in two essential parts: it ignores the model-internal representation employed for reconstruction, and it lacks formal assertions and comparability between samples. We address these shortcomings by proposing the Context-encoding Variational Autoencoder (ceVAE) which combines reconstruction- with density-based anomaly scoring. This improves the sample- as well as pixel-wise results. In our experiments on the BraTS-2017 and ISLES-2015 segmentation benchmarks, the ceVAE achieves unsupervised ROC-AUCs of 0.95 and 0.89, respectively, thus outperforming state-of-the-art methods by a considerable margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge