Chengfeng Zhou
OphthBench: A Comprehensive Benchmark for Evaluating Large Language Models in Chinese Ophthalmology
Feb 03, 2025Abstract:Large language models (LLMs) have shown significant promise across various medical applications, with ophthalmology being a notable area of focus. Many ophthalmic tasks have shown substantial improvement through the integration of LLMs. However, before these models can be widely adopted in clinical practice, evaluating their capabilities and identifying their limitations is crucial. To address this research gap and support the real-world application of LLMs, we introduce the OphthBench, a specialized benchmark designed to assess LLM performance within the context of Chinese ophthalmic practices. This benchmark systematically divides a typical ophthalmic clinical workflow into five key scenarios: Education, Triage, Diagnosis, Treatment, and Prognosis. For each scenario, we developed multiple tasks featuring diverse question types, resulting in a comprehensive benchmark comprising 9 tasks and 591 questions. This comprehensive framework allows for a thorough assessment of LLMs' capabilities and provides insights into their practical application in Chinese ophthalmology. Using this benchmark, we conducted extensive experiments and analyzed the results from 39 popular LLMs. Our evaluation highlights the current gap between LLM development and its practical utility in clinical settings, providing a clear direction for future advancements. By bridging this gap, we aim to unlock the potential of LLMs and advance their development in ophthalmology.
SPRMamba: Surgical Phase Recognition for Endoscopic Submucosal Dissection with Mamba
Sep 18, 2024



Abstract:Endoscopic Submucosal Dissection (ESD) is a minimally invasive procedure initially designed for the treatment of early gastric cancer but is now widely used for various gastrointestinal lesions. Computer-assisted Surgery systems have played a crucial role in improving the precision and safety of ESD procedures, however, their effectiveness is limited by the accurate recognition of surgical phases. The intricate nature of ESD, with different lesion characteristics and tissue structures, presents challenges for real-time surgical phase recognition algorithms. Existing surgical phase recognition algorithms struggle to efficiently capture temporal contexts in video-based scenarios, leading to insufficient performance. To address these issues, we propose SPRMamba, a novel Mamba-based framework for ESD surgical phase recognition. SPRMamba leverages the strengths of Mamba for long-term temporal modeling while introducing the Scaled Residual TranMamba block to enhance the capture of fine-grained details, overcoming the limitations of traditional temporal models like Temporal Convolutional Networks and Transformers. Moreover, a Temporal Sample Strategy is introduced to accelerate the processing, which is essential for real-time phase recognition in clinical settings. Extensive testing on the ESD385 dataset and the cholecystectomy Cholec80 dataset demonstrates that SPRMamba surpasses existing state-of-the-art methods and exhibits greater robustness across various surgical phase recognition tasks.
PredIN: Towards Open-Set Gesture Recognition via Prediction Inconsistency
Jul 29, 2024Abstract:Gesture recognition based on surface electromyography (sEMG) has achieved significant progress in human-machine interaction (HMI). However, accurately recognizing predefined gestures within a closed set is still inadequate in practice; a robust open-set system needs to effectively reject unknown gestures while correctly classifying known ones. To handle this challenge, we first report prediction inconsistency discovered for unknown classes due to ensemble diversity, which can significantly facilitate the detection of unknown classes. Based on this insight, we propose an ensemble learning approach, PredIN, to explicitly magnify the prediction inconsistency by enhancing ensemble diversity. Specifically, PredIN maximizes the class feature distribution inconsistency among ensemble members to enhance diversity. Meanwhile, it optimizes inter-class separability within an individual ensemble member to maintain individual performance. Comprehensive experiments on various benchmark datasets demonstrate that the PredIN outperforms state-of-the-art methods by a clear margin.Our proposed method simultaneously achieves accurate closed-set classification for predefined gestures and effective rejection for unknown gestures, exhibiting its efficacy and superiority in open-set gesture recognition based on sEMG.
Skeleton-Guided Instance Separation for Fine-Grained Segmentation in Microscopy
Jan 19, 2024



Abstract:One of the fundamental challenges in microscopy (MS) image analysis is instance segmentation (IS), particularly when segmenting cluster regions where multiple objects of varying sizes and shapes may be connected or even overlapped in arbitrary orientations. Existing IS methods usually fail in handling such scenarios, as they rely on coarse instance representations such as keypoints and horizontal bounding boxes (h-bboxes). In this paper, we propose a novel one-stage framework named A2B-IS to address this challenge and enhance the accuracy of IS in MS images. Our approach represents each instance with a pixel-level mask map and a rotated bounding box (r-bbox). Unlike two-stage methods that use box proposals for segmentations, our method decouples mask and box predictions, enabling simultaneous processing to streamline the model pipeline. Additionally, we introduce a Gaussian skeleton map to aid the IS task in two key ways: (1) It guides anchor placement, reducing computational costs while improving the model's capacity to learn RoI-aware features by filtering out noise from background regions. (2) It ensures accurate isolation of densely packed instances by rectifying erroneous box predictions near instance boundaries. To further enhance the performance, we integrate two modules into the framework: (1) An Atrous Attention Block (A2B) designed to extract high-resolution feature maps with fine-grained multiscale information, and (2) A Semi-Supervised Learning (SSL) strategy that leverages both labeled and unlabeled images for model training. Our method has been thoroughly validated on two large-scale MS datasets, demonstrating its superiority over most state-of-the-art approaches.
Towards Open-set Gesture Recognition via Feature Activation Enhancement and Orthogonal Prototype Learning
Dec 05, 2023Abstract:Gesture recognition is a foundational task in human-machine interaction (HMI). While there has been significant progress in gesture recognition based on surface electromyography (sEMG), accurate recognition of predefined gestures only within a closed set is still inadequate in practice. It is essential to effectively discern and reject unknown gestures of disinterest in a robust system. Numerous methods based on prototype learning (PL) have been proposed to tackle this open set recognition (OSR) problem. However, they do not fully explore the inherent distinctions between known and unknown classes. In this paper, we propose a more effective PL method leveraging two novel and inherent distinctions, feature activation level and projection inconsistency. Specifically, the Feature Activation Enhancement Mechanism (FAEM) widens the gap in feature activation values between known and unknown classes. Furthermore, we introduce Orthogonal Prototype Learning (OPL) to construct multiple perspectives. OPL acts to project a sample from orthogonal directions to maximize the distinction between its two projections, where unknown samples will be projected near the clusters of different known classes while known samples still maintain intra-class similarity. Our proposed method simultaneously achieves accurate closed-set classification for predefined gestures and effective rejection for unknown gestures. Extensive experiments demonstrate its efficacy and superiority in open-set gesture recognition based on sEMG.
Towards Discriminative Representation with Meta-learning for Colonoscopic Polyp Re-Identification
Aug 02, 2023



Abstract:Colonoscopic Polyp Re-Identification aims to match the same polyp from a large gallery with images from different views taken using different cameras and plays an important role in the prevention and treatment of colorectal cancer in computer-aided diagnosis. However, traditional methods for object ReID directly adopting CNN models trained on the ImageNet dataset usually produce unsatisfactory retrieval performance on colonoscopic datasets due to the large domain gap. Additionally, these methods neglect to explore the potential of self-discrepancy among intra-class relations in the colonoscopic polyp dataset, which remains an open research problem in the medical community. To solve this dilemma, we propose a simple but effective training method named Colo-ReID, which can help our model to learn more general and discriminative knowledge based on the meta-learning strategy in scenarios with fewer samples. Based on this, a dynamic Meta-Learning Regulation mechanism called MLR is introduced to further boost the performance of polyp re-identification. To the best of our knowledge, this is the first attempt to leverage the meta-learning paradigm instead of traditional machine learning to effectively train deep models in the task of colonoscopic polyp re-identification. Empirical results show that our method significantly outperforms current state-of-the-art methods by a clear margin.
Learning Robust Visual-Semantic Embedding for Generalizable Person Re-identification
Apr 19, 2023Abstract:Generalizable person re-identification (Re-ID) is a very hot research topic in machine learning and computer vision, which plays a significant role in realistic scenarios due to its various applications in public security and video surveillance. However, previous methods mainly focus on the visual representation learning, while neglect to explore the potential of semantic features during training, which easily leads to poor generalization capability when adapted to the new domain. In this paper, we propose a Multi-Modal Equivalent Transformer called MMET for more robust visual-semantic embedding learning on visual, textual and visual-textual tasks respectively. To further enhance the robust feature learning in the context of transformer, a dynamic masking mechanism called Masked Multimodal Modeling strategy (MMM) is introduced to mask both the image patches and the text tokens, which can jointly works on multimodal or unimodal data and significantly boost the performance of generalizable person Re-ID. Extensive experiments on benchmark datasets demonstrate the competitive performance of our method over previous approaches. We hope this method could advance the research towards visual-semantic representation learning. Our source code is also publicly available at https://github.com/JeremyXSC/MMET.
A simple normalization technique using window statistics to improve the out-of-distribution generalization on medical images
Jul 14, 2022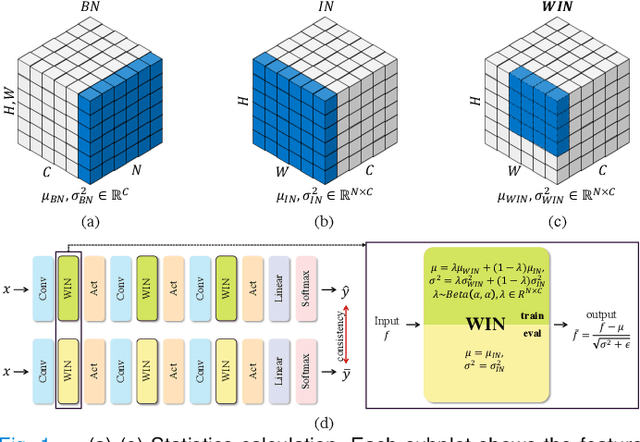
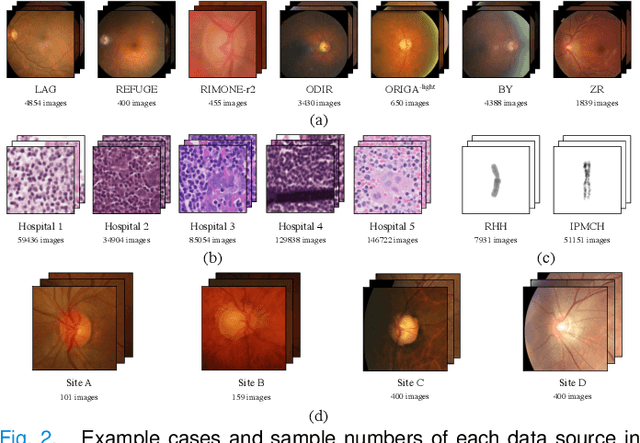
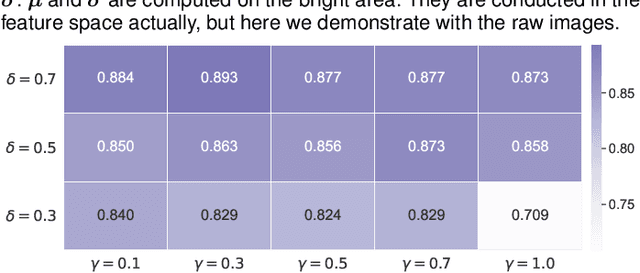
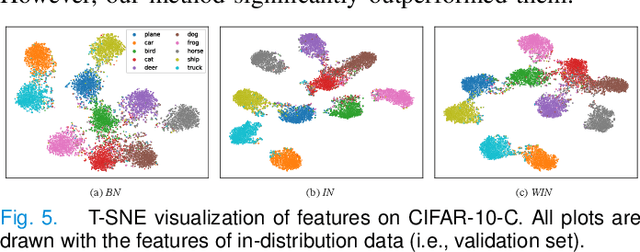
Abstract:Since data scarcity and data heterogeneity are prevailing for medical images, well-trained Convolutional Neural Networks (CNNs) using previous normalization methods may perform poorly when deployed to a new site. However, a reliable model for real-world clinical applications should be able to generalize well both on in-distribution (IND) and out-of-distribution (OOD) data (e.g., the new site data). In this study, we present a novel normalization technique called window normalization (WIN) to improve the model generalization on heterogeneous medical images, which is a simple yet effective alternative to existing normalization methods. Specifically, WIN perturbs the normalizing statistics with the local statistics computed on the window of features. This feature-level augmentation technique regularizes the models well and improves their OOD generalization significantly. Taking its advantage, we propose a novel self-distillation method called WIN-WIN for classification tasks. WIN-WIN is easily implemented with twice forward passes and a consistency constraint, which can be a simple extension for existing methods. Extensive experimental results on various tasks (6 tasks) and datasets (24 datasets) demonstrate the generality and effectiveness of our methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge