Brian Davidson
Competing for pixels: a self-play algorithm for weakly-supervised segmentation
May 26, 2024Abstract:Weakly-supervised segmentation (WSS) methods, reliant on image-level labels indicating object presence, lack explicit correspondence between labels and regions of interest (ROIs), posing a significant challenge. Despite this, WSS methods have attracted attention due to their much lower annotation costs compared to fully-supervised segmentation. Leveraging reinforcement learning (RL) self-play, we propose a novel WSS method that gamifies image segmentation of a ROI. We formulate segmentation as a competition between two agents that compete to select ROI-containing patches until exhaustion of all such patches. The score at each time-step, used to compute the reward for agent training, represents likelihood of object presence within the selection, determined by an object presence detector pre-trained using only image-level binary classification labels of object presence. Additionally, we propose a game termination condition that can be called by either side upon exhaustion of all ROI-containing patches, followed by the selection of a final patch from each. Upon termination, the agent is incentivised if ROI-containing patches are exhausted or disincentivised if an ROI-containing patch is found by the competitor. This competitive setup ensures minimisation of over- or under-segmentation, a common problem with WSS methods. Extensive experimentation across four datasets demonstrates significant performance improvements over recent state-of-the-art methods. Code: https://github.com/s-sd/spurl/tree/main/wss
Active learning using adaptable task-based prioritisation
Dec 03, 2022



Abstract:Supervised machine learning-based medical image computing applications necessitate expert label curation, while unlabelled image data might be relatively abundant. Active learning methods aim to prioritise a subset of available image data for expert annotation, for label-efficient model training. We develop a controller neural network that measures priority of images in a sequence of batches, as in batch-mode active learning, for multi-class segmentation tasks. The controller is optimised by rewarding positive task-specific performance gain, within a Markov decision process (MDP) environment that also optimises the task predictor. In this work, the task predictor is a segmentation network. A meta-reinforcement learning algorithm is proposed with multiple MDPs, such that the pre-trained controller can be adapted to a new MDP that contains data from different institutes and/or requires segmentation of different organs or structures within the abdomen. We present experimental results using multiple CT datasets from more than one thousand patients, with segmentation tasks of nine different abdominal organs, to demonstrate the efficacy of the learnt prioritisation controller function and its cross-institute and cross-organ adaptability. We show that the proposed adaptable prioritisation metric yields converging segmentation accuracy for the novel class of kidney, unseen in training, using between approximately 40\% to 60\% of labels otherwise required with other heuristic or random prioritisation metrics. For clinical datasets of limited size, the proposed adaptable prioritisation offers a performance improvement of 22.6\% and 10.2\% in Dice score, for tasks of kidney and liver vessel segmentation, respectively, compared to random prioritisation and alternative active sampling strategies.
Voice-assisted Image Labelling for Endoscopic Ultrasound Classification using Neural Networks
Oct 12, 2021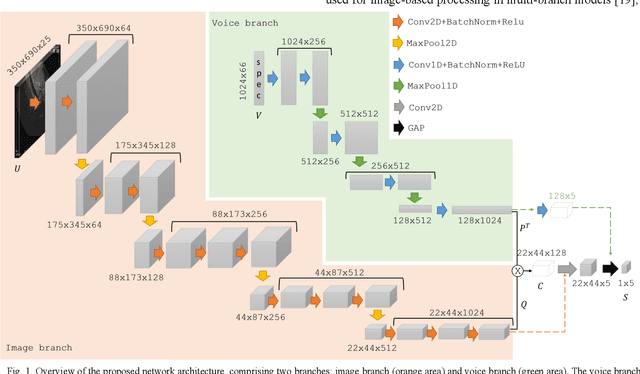
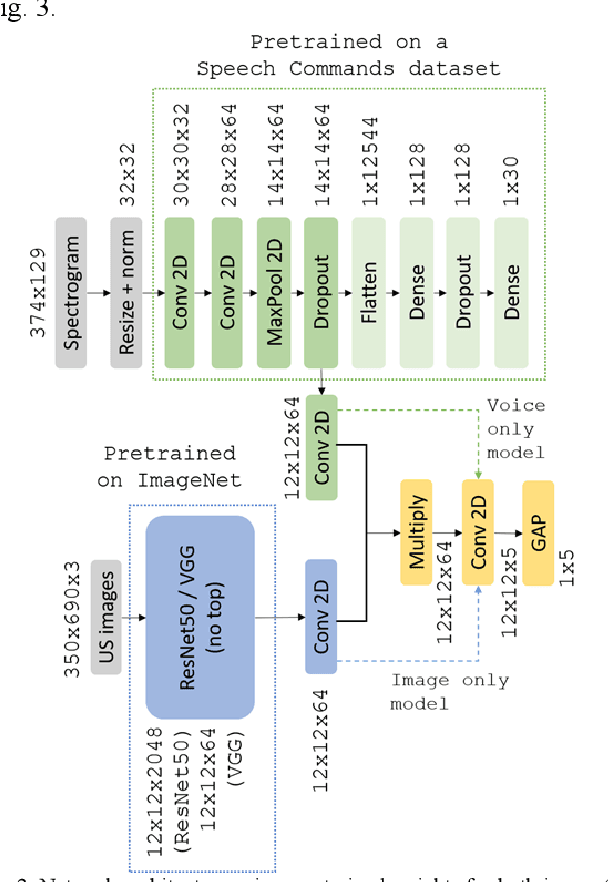
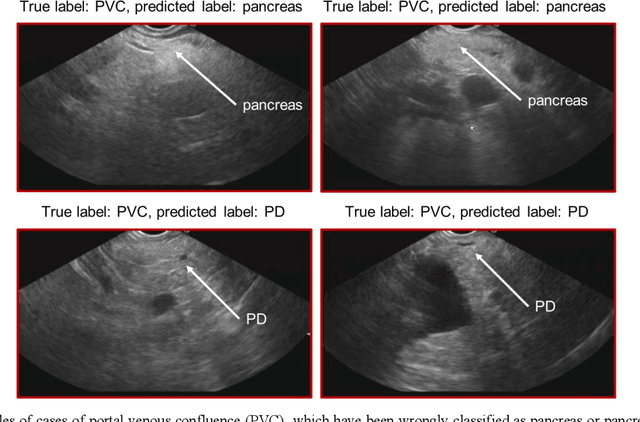
Abstract:Ultrasound imaging is a commonly used technology for visualising patient anatomy in real-time during diagnostic and therapeutic procedures. High operator dependency and low reproducibility make ultrasound imaging and interpretation challenging with a steep learning curve. Automatic image classification using deep learning has the potential to overcome some of these challenges by supporting ultrasound training in novices, as well as aiding ultrasound image interpretation in patient with complex pathology for more experienced practitioners. However, the use of deep learning methods requires a large amount of data in order to provide accurate results. Labelling large ultrasound datasets is a challenging task because labels are retrospectively assigned to 2D images without the 3D spatial context available in vivo or that would be inferred while visually tracking structures between frames during the procedure. In this work, we propose a multi-modal convolutional neural network (CNN) architecture that labels endoscopic ultrasound (EUS) images from raw verbal comments provided by a clinician during the procedure. We use a CNN composed of two branches, one for voice data and another for image data, which are joined to predict image labels from the spoken names of anatomical landmarks. The network was trained using recorded verbal comments from expert operators. Our results show a prediction accuracy of 76% at image level on a dataset with 5 different labels. We conclude that the addition of spoken commentaries can increase the performance of ultrasound image classification, and eliminate the burden of manually labelling large EUS datasets necessary for deep learning applications.
Intraoperative Liver Surface Completion with Graph Convolutional VAE
Sep 08, 2020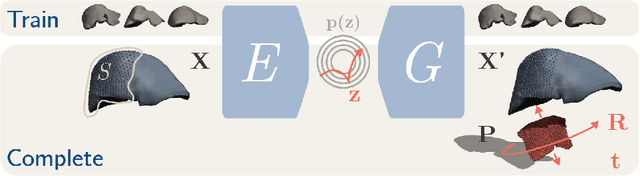
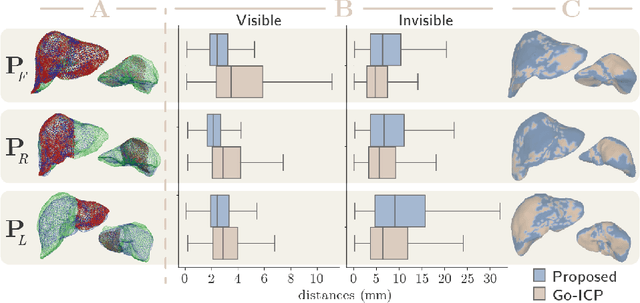
Abstract:In this work we propose a method based on geometric deep learning to predict the complete surface of the liver, given a partial point cloud of the organ obtained during the surgical laparoscopic procedure. We introduce a new data augmentation technique that randomly perturbs shapes in their frequency domain to compensate the limited size of our dataset. The core of our method is a variational autoencoder (VAE) that is trained to learn a latent space for complete shapes of the liver. At inference time, the generative part of the model is embedded in an optimisation procedure where the latent representation is iteratively updated to generate a model that matches the intraoperative partial point cloud. The effect of this optimisation is a progressive non-rigid deformation of the initially generated shape. Our method is qualitatively evaluated on real data and quantitatively evaluated on synthetic data. We compared with a state-of-the-art rigid registration algorithm, that our method outperformed in visible areas.
More unlabelled data or label more data? A study on semi-supervised laparoscopic image segmentation
Aug 20, 2019
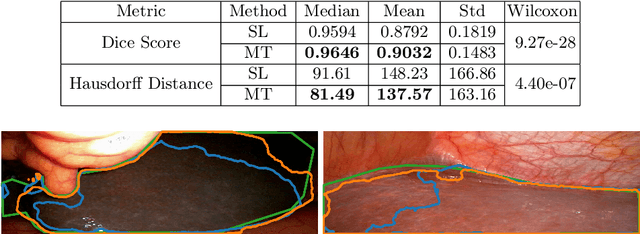
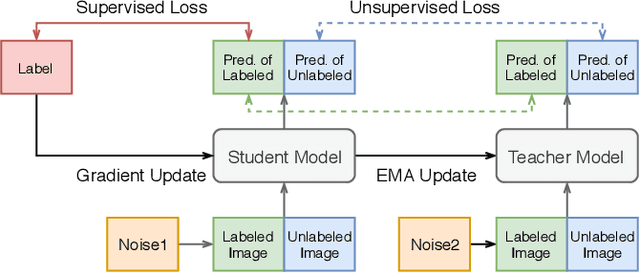
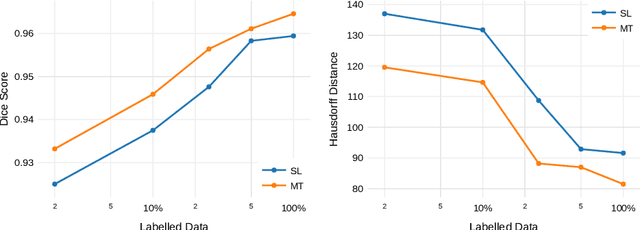
Abstract:Improving a semi-supervised image segmentation task has the option of adding more unlabelled images, labelling the unlabelled images or combining both, as neither image acquisition nor expert labelling can be considered trivial in most clinical applications. With a laparoscopic liver image segmentation application, we investigate the performance impact by altering the quantities of labelled and unlabelled training data, using a semi-supervised segmentation algorithm based on the mean teacher learning paradigm. We first report a significantly higher segmentation accuracy, compared with supervised learning. Interestingly, this comparison reveals that the training strategy adopted in the semi-supervised algorithm is also responsible for this observed improvement, in addition to the added unlabelled data. We then compare different combinations of labelled and unlabelled data set sizes for training semi-supervised segmentation networks, to provide a quantitative example of the practically useful trade-off between the two data planning strategies in this surgical guidance application.
Whole-Sample Mapping of Cancerous and Benign Tissue Properties
Jul 23, 2019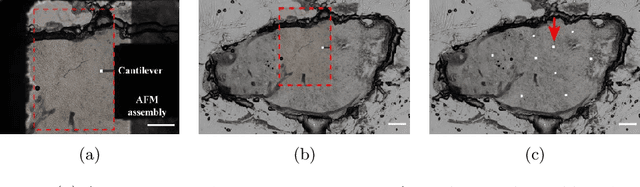
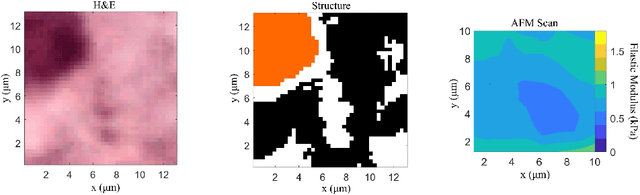

Abstract:Structural and mechanical differences between cancerous and healthy tissue give rise to variations in macroscopic properties such as visual appearance and elastic modulus that show promise as signatures for early cancer detection. Atomic force microscopy (AFM) has been used to measure significant differences in stiffness between cancerous and healthy cells owing to its high force sensitivity and spatial resolution, however due to absorption and scattering of light, it is often challenging to accurately locate where AFM measurements have been made on a bulk tissue sample. In this paper we describe an image registration method that localizes AFM elastic stiffness measurements with high-resolution images of haematoxylin and eosin (H\&E)-stained tissue to within 1.5 microns. Color RGB images are segmented into three structure types (lumen, cells and stroma) by a neural network classifier trained on ground-truth pixel data obtained through k-means clustering in HSV color space. Using the localized stiffness maps and corresponding structural information, a whole-sample stiffness map is generated with a region matching and interpolation algorithm that associates similar structures with measured stiffness values. We present results showing significant differences in stiffness between healthy and cancerous liver tissue and discuss potential applications of this technique.
Augmented Reality needle ablation guidance tool for Irreversible Electroporation in the pancreas
Feb 09, 2018Abstract:Irreversible electroporation (IRE) is a soft tissue ablation technique suitable for treatment of inoperable tumours in the pancreas. The process involves applying a high voltage electric field to the tissue containing the mass using needle electrodes, leaving cancerous cells irreversibly damaged and vulnerable to apoptosis. Efficacy of the treatment depends heavily on the accuracy of needle placement and requires a high degree of skill from the operator. In this paper, we describe an Augmented Reality (AR) system designed to overcome the challenges associated with planning and guiding the needle insertion process. Our solution, based on the HoloLens (Microsoft, USA) platform, tracks the position of the headset, needle electrodes and ultrasound (US) probe in space. The proof of concept implementation of the system uses this tracking data to render real-time holographic guides on the HoloLens, giving the user insight into the current progress of needle insertion and an indication of the target needle trajectory. The operator's field of view is augmented using visual guides and real-time US feed rendered on a holographic plane, eliminating the need to consult external monitors. Based on these early prototypes, we are aiming to develop a system that will lower the skill level required for IRE while increasing overall accuracy of needle insertion and, hence, the likelihood of successful treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge