Alexandre Gramfort
MIND - INRIA
Averaging Spatio-temporal Signals using Optimal Transport and Soft Alignments
Apr 08, 2022Abstract:Several fields in science, from genomics to neuroimaging, require monitoring populations (measures) that evolve with time. These complex datasets, describing dynamics with both time and spatial components, pose new challenges for data analysis. We propose in this work a new framework to carry out averaging of these datasets, with the goal of synthesizing a representative template trajectory from multiple trajectories. We show that this requires addressing three sources of invariance: shifts in time, space, and total population size (or mass/amplitude). Here we draw inspiration from dynamic time warping (DTW), optimal transport (OT) theory and its unbalanced extension (UOT) to propose a criterion that can address all three issues. This proposal leverages a smooth formulation of DTW (Soft-DTW) that is shown to capture temporal shifts, and UOT to handle both variations in space and size. Our proposed loss can be used to define spatio-temporal barycenters as Fr\'echet means. Using Fenchel duality, we show how these barycenters can be computed efficiently, in parallel, via a novel variant of entropy-regularized debiased UOT. Experiments on handwritten letters and brain imaging data confirm our theoretical findings and illustrate the effectiveness of the proposed loss for spatio-temporal data.
The Optimal Noise in Noise-Contrastive Learning Is Not What You Think
Mar 02, 2022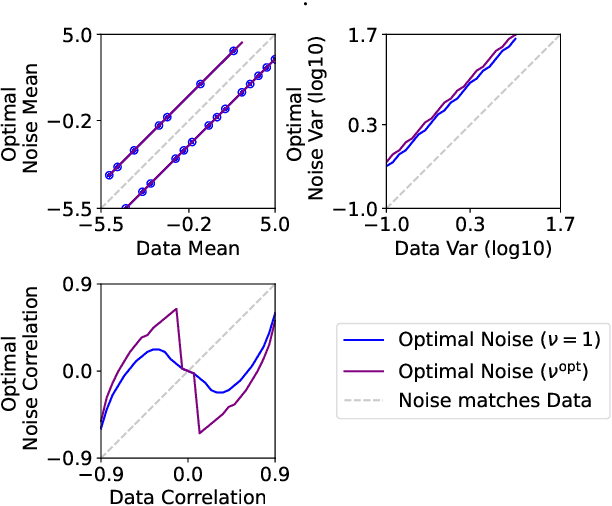
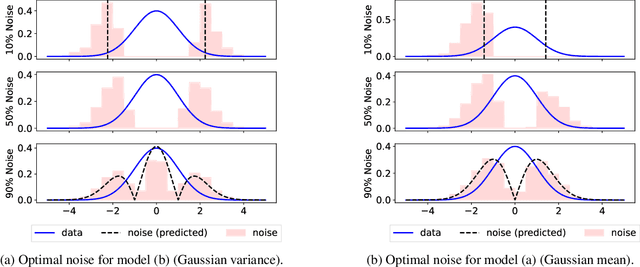
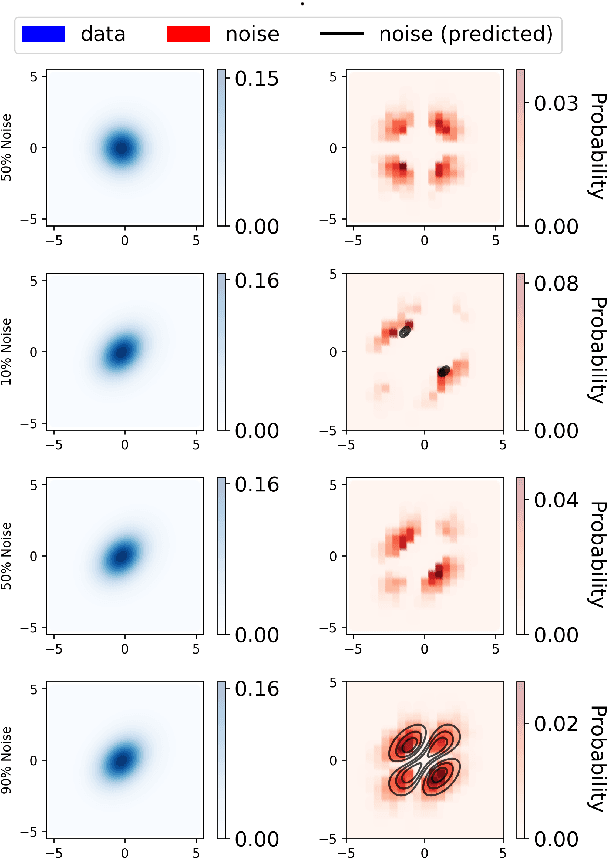
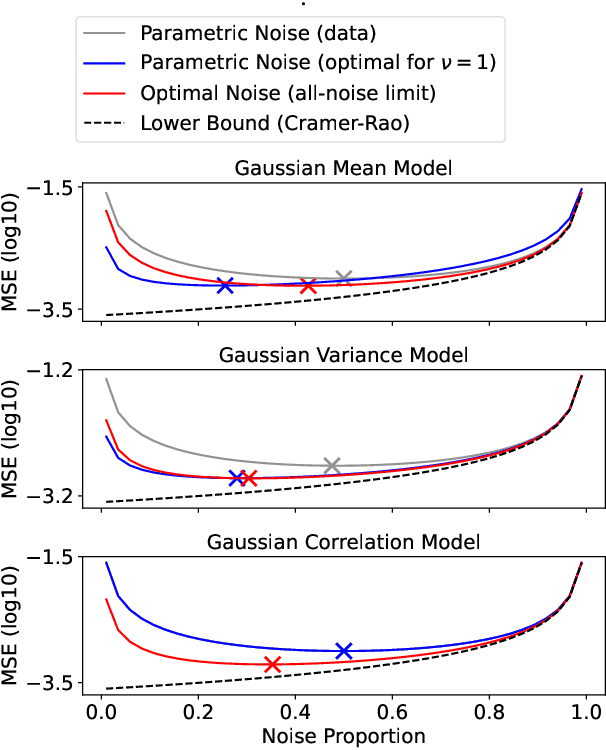
Abstract:Learning a parametric model of a data distribution is a well-known statistical problem that has seen renewed interest as it is brought to scale in deep learning. Framing the problem as a self-supervised task, where data samples are discriminated from noise samples, is at the core of state-of-the-art methods, beginning with Noise-Contrastive Estimation (NCE). Yet, such contrastive learning requires a good noise distribution, which is hard to specify; domain-specific heuristics are therefore widely used. While a comprehensive theory is missing, it is widely assumed that the optimal noise should in practice be made equal to the data, both in distribution and proportion. This setting underlies Generative Adversarial Networks (GANs) in particular. Here, we empirically and theoretically challenge this assumption on the optimal noise. We show that deviating from this assumption can actually lead to better statistical estimators, in terms of asymptotic variance. In particular, the optimal noise distribution is different from the data's and even from a different family.
Deep invariant networks with differentiable augmentation layers
Feb 16, 2022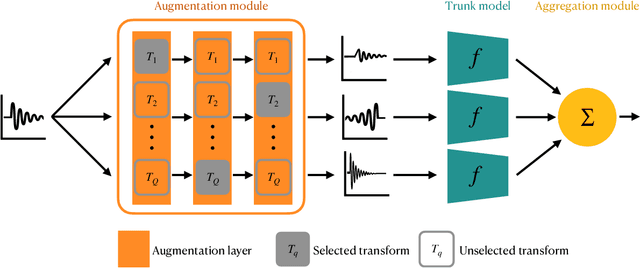
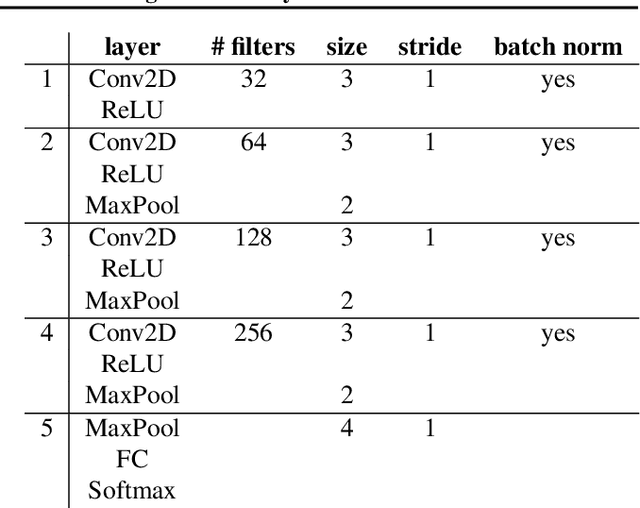

Abstract:Designing learning systems which are invariant to certain data transformations is critical in machine learning. Practitioners can typically enforce a desired invariance on the trained model through the choice of a network architecture, e.g. using convolutions for translations, or using data augmentation. Yet, enforcing true invariance in the network can be difficult, and data invariances are not always known a piori. State-of-the-art methods for learning data augmentation policies require held-out data and are based on bilevel optimization problems, which are complex to solve and often computationally demanding. In this work we investigate new ways of learning invariances only from the training data. Using learnable augmentation layers built directly in the network, we demonstrate that our method is very versatile. It can incorporate any type of differentiable augmentation and be applied to a broad class of learning problems beyond computer vision. We provide empirical evidence showing that our approach is easier and faster to train than modern automatic data augmentation techniques based on bilevel optimization, while achieving comparable results. Experiments show that while the invariances transferred to a model through automatic data augmentation are limited by the model expressivity, the invariance yielded by our approach is insensitive to it by design.
2021 BEETL Competition: Advancing Transfer Learning for Subject Independence & Heterogenous EEG Data Sets
Feb 14, 2022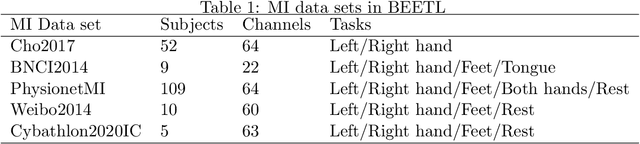
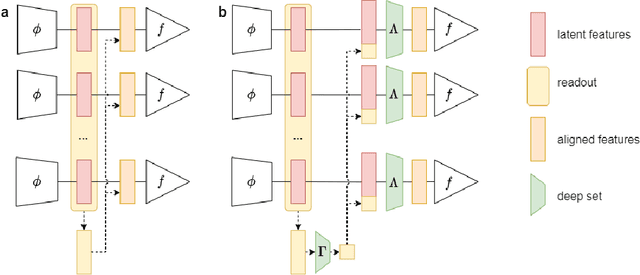
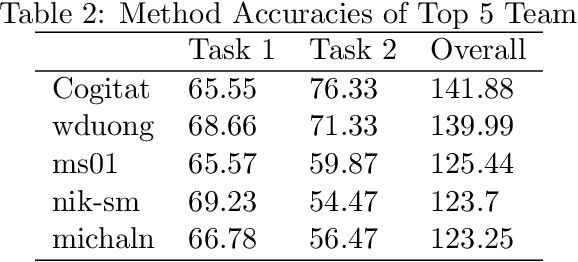
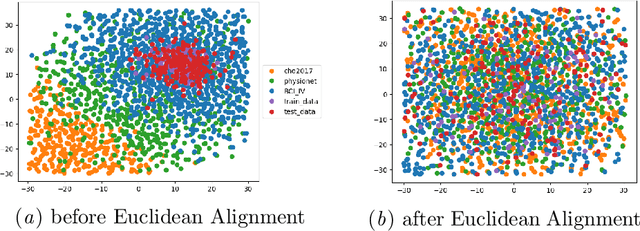
Abstract:Transfer learning and meta-learning offer some of the most promising avenues to unlock the scalability of healthcare and consumer technologies driven by biosignal data. This is because current methods cannot generalise well across human subjects' data and handle learning from different heterogeneously collected data sets, thus limiting the scale of training data. On the other side, developments in transfer learning would benefit significantly from a real-world benchmark with immediate practical application. Therefore, we pick electroencephalography (EEG) as an exemplar for what makes biosignal machine learning hard. We design two transfer learning challenges around diagnostics and Brain-Computer-Interfacing (BCI), that have to be solved in the face of low signal-to-noise ratios, major variability among subjects, differences in the data recording sessions and techniques, and even between the specific BCI tasks recorded in the dataset. Task 1 is centred on the field of medical diagnostics, addressing automatic sleep stage annotation across subjects. Task 2 is centred on Brain-Computer Interfacing (BCI), addressing motor imagery decoding across both subjects and data sets. The BEETL competition with its over 30 competing teams and its 3 winning entries brought attention to the potential of deep transfer learning and combinations of set theory and conventional machine learning techniques to overcome the challenges. The results set a new state-of-the-art for the real-world BEETL benchmark.
DriPP: Driven Point Processes to Model Stimuli Induced Patterns in M/EEG Signals
Dec 08, 2021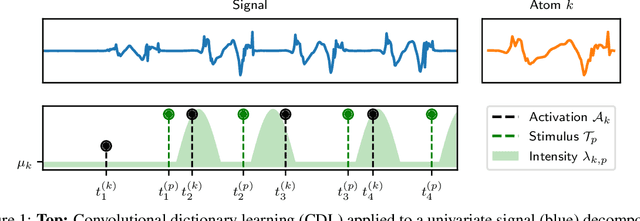
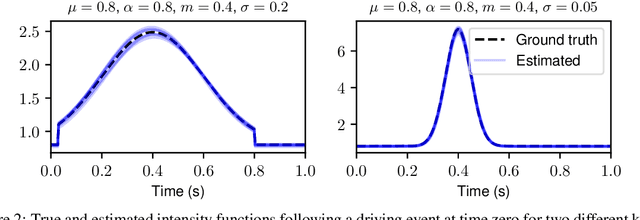
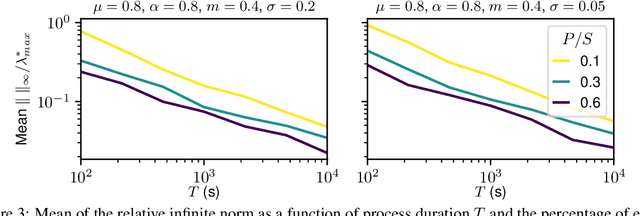
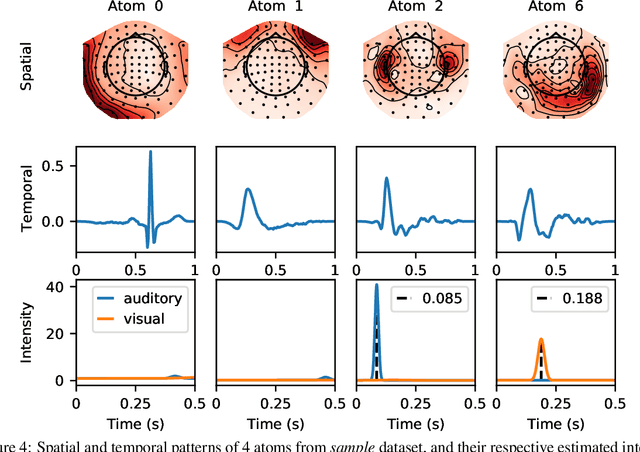
Abstract:The quantitative analysis of non-invasive electrophysiology signals from electroencephalography (EEG) and magnetoencephalography (MEG) boils down to the identification of temporal patterns such as evoked responses, transient bursts of neural oscillations but also blinks or heartbeats for data cleaning. Several works have shown that these patterns can be extracted efficiently in an unsupervised way, e.g., using Convolutional Dictionary Learning. This leads to an event-based description of the data. Given these events, a natural question is to estimate how their occurrences are modulated by certain cognitive tasks and experimental manipulations. To address it, we propose a point process approach. While point processes have been used in neuroscience in the past, in particular for single cell recordings (spike trains), techniques such as Convolutional Dictionary Learning make them amenable to human studies based on EEG/MEG signals. We develop a novel statistical point process model-called driven temporal point processes (DriPP)-where the intensity function of the point process model is linked to a set of point processes corresponding to stimulation events. We derive a fast and principled expectation-maximization (EM) algorithm to estimate the parameters of this model. Simulations reveal that model parameters can be identified from long enough signals. Results on standard MEG datasets demonstrate that our methodology reveals event-related neural responses-both evoked and induced-and isolates non-task specific temporal patterns.
Long-range and hierarchical language predictions in brains and algorithms
Nov 28, 2021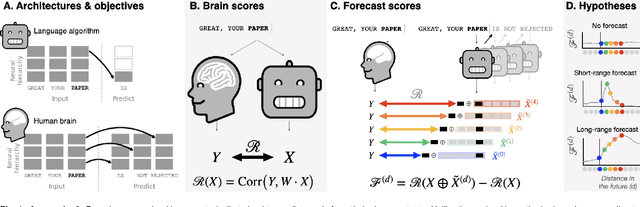
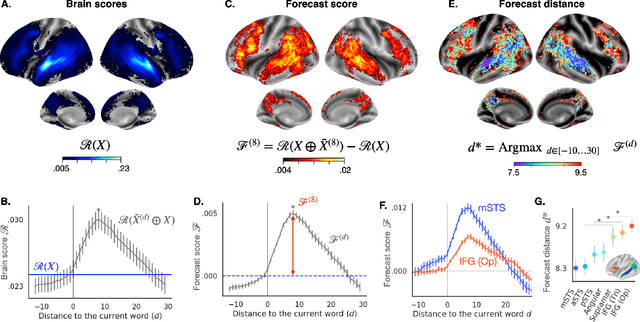
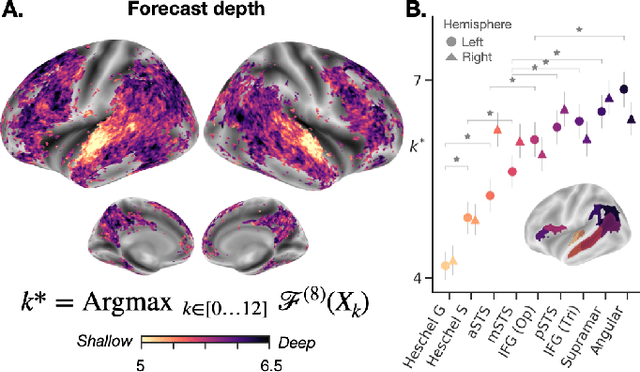
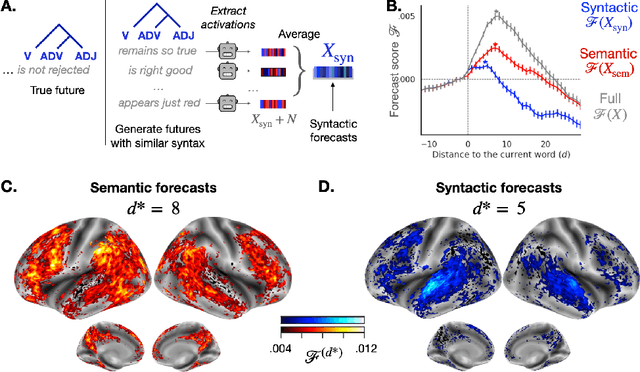
Abstract:Deep learning has recently made remarkable progress in natural language processing. Yet, the resulting algorithms remain far from competing with the language abilities of the human brain. Predictive coding theory offers a potential explanation to this discrepancy: while deep language algorithms are optimized to predict adjacent words, the human brain would be tuned to make long-range and hierarchical predictions. To test this hypothesis, we analyze the fMRI brain signals of 304 subjects each listening to 70min of short stories. After confirming that the activations of deep language algorithms linearly map onto those of the brain, we show that enhancing these models with long-range forecast representations improves their brain-mapping. The results further reveal a hierarchy of predictions in the brain, whereby the fronto-parietal cortices forecast more abstract and more distant representations than the temporal cortices. Overall, this study strengthens predictive coding theory and suggests a critical role of long-range and hierarchical predictions in natural language processing.
Inverting brain grey matter models with likelihood-free inference: a tool for trustable cytoarchitecture measurements
Nov 15, 2021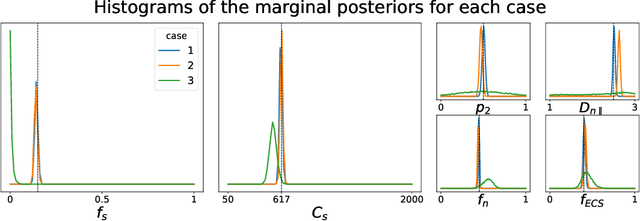
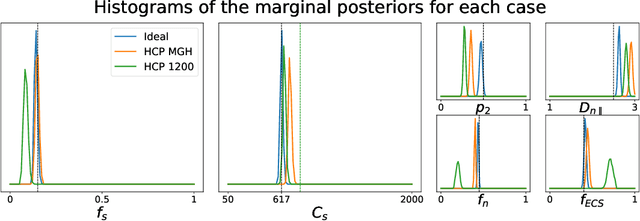
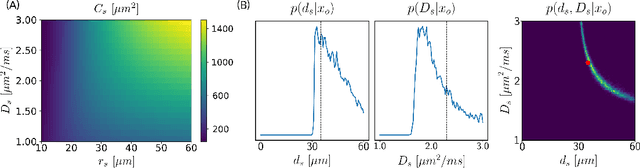
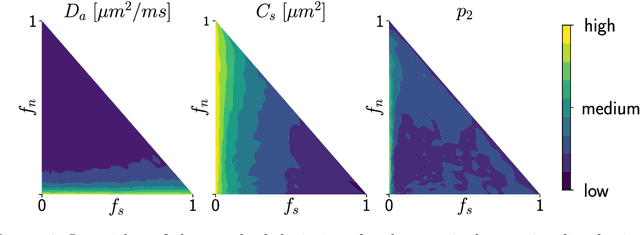
Abstract:Effective characterisation of the brain grey matter cytoarchitecture with quantitative sensitivity to soma density and volume remains an unsolved challenge in diffusion MRI (dMRI). Solving the problem of relating the dMRI signal with cytoarchitectural characteristics calls for the definition of a mathematical model that describes brain tissue via a handful of physiologically-relevant parameters and an algorithm for inverting the model. To address this issue, we propose a new forward model, specifically a new system of equations, requiring a few relatively sparse b-shells. We then apply modern tools from Bayesian analysis known as likelihood-free inference (LFI) to invert our proposed model. As opposed to other approaches from the literature, our algorithm yields not only an estimation of the parameter vector $\theta$ that best describes a given observed data point $x_0$, but also a full posterior distribution $p(\theta|x_0)$ over the parameter space. This enables a richer description of the model inversion, providing indicators such as credible intervals for the estimated parameters and a complete characterization of the parameter regions where the model may present indeterminacies. We approximate the posterior distribution using deep neural density estimators, known as normalizing flows, and fit them using a set of repeated simulations from the forward model. We validate our approach on simulations using dmipy and then apply the whole pipeline on two publicly available datasets.
LassoBench: A High-Dimensional Hyperparameter Optimization Benchmark Suite for Lasso
Nov 04, 2021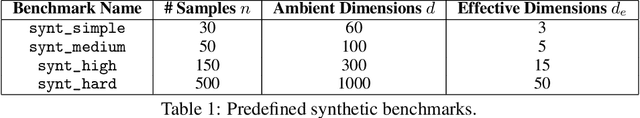
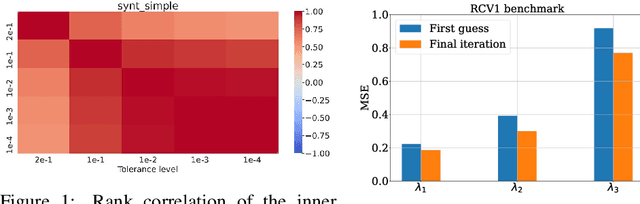

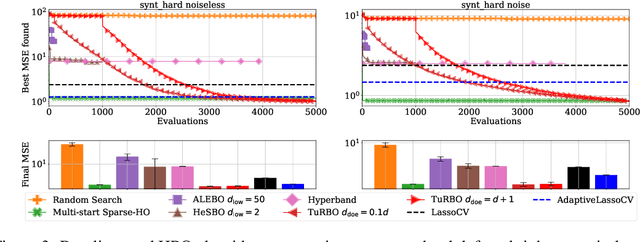
Abstract:Even though Weighted Lasso regression has appealing statistical guarantees, it is typically avoided due to its complex search space described with thousands of hyperparameters. On the other hand, the latest progress with high-dimensional HPO methods for black-box functions demonstrates that high-dimensional applications can indeed be efficiently optimized. Despite this initial success, the high-dimensional HPO approaches are typically applied to synthetic problems with a moderate number of dimensions which limits its impact in scientific and engineering applications. To address this limitation, we propose LassoBench, a new benchmark suite tailored for an important open research topic in the Lasso community that is Weighted Lasso regression. LassoBench consists of benchmarks on both well-controlled synthetic setups (number of samples, SNR, ambient and effective dimensionalities, and multiple fidelities) and real-world datasets, which enable the use of many flavors of HPO algorithms to be improved and extended to the high-dimensional setting. We evaluate 5 state-of-the-art HPO methods and 3 baselines, and demonstrate that Bayesian optimization, in particular, can improve over the methods commonly used for sparse regression while highlighting limitations of these frameworks in very high-dimensions. Remarkably, Bayesian optimization improve the Lasso baselines on 60, 100, 300, and 1000 dimensional problems by 45.7%, 19.2%, 19.7% and 15.5%, respectively.
Shared Independent Component Analysis for Multi-Subject Neuroimaging
Oct 26, 2021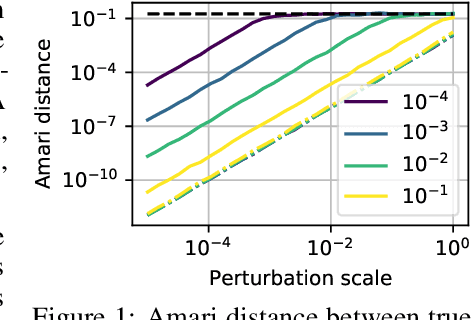
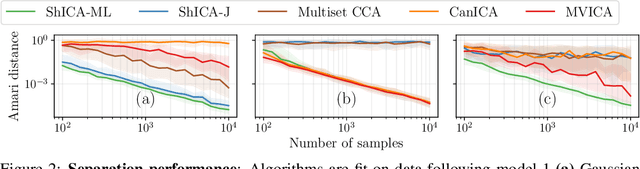
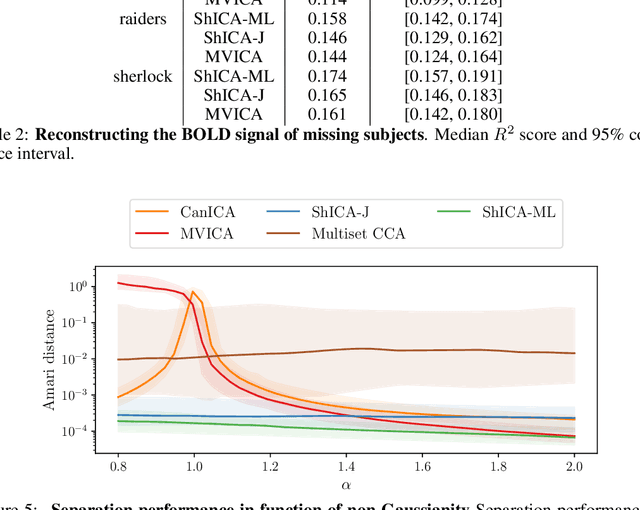
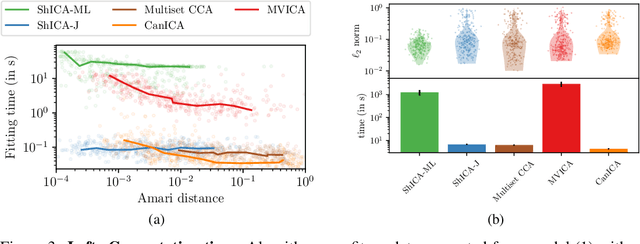
Abstract:We consider shared response modeling, a multi-view learning problem where one wants to identify common components from multiple datasets or views. We introduce Shared Independent Component Analysis (ShICA) that models each view as a linear transform of shared independent components contaminated by additive Gaussian noise. We show that this model is identifiable if the components are either non-Gaussian or have enough diversity in noise variances. We then show that in some cases multi-set canonical correlation analysis can recover the correct unmixing matrices, but that even a small amount of sampling noise makes Multiset CCA fail. To solve this problem, we propose to use joint diagonalization after Multiset CCA, leading to a new approach called ShICA-J. We show via simulations that ShICA-J leads to improved results while being very fast to fit. While ShICA-J is based on second-order statistics, we further propose to leverage non-Gaussianity of the components using a maximum-likelihood method, ShICA-ML, that is both more accurate and more costly. Further, ShICA comes with a principled method for shared components estimation. Finally, we provide empirical evidence on fMRI and MEG datasets that ShICA yields more accurate estimation of the components than alternatives.
Label scarcity in biomedicine: Data-rich latent factor discovery enhances phenotype prediction
Oct 12, 2021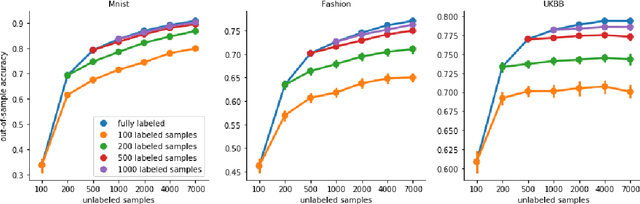
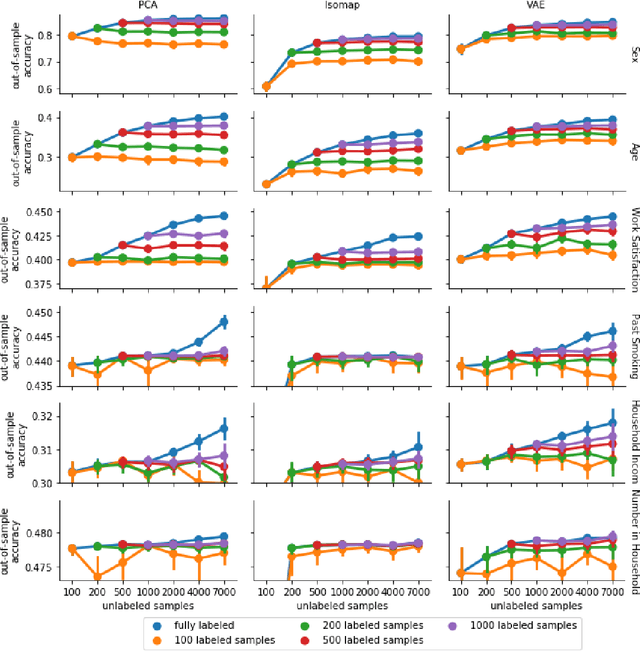
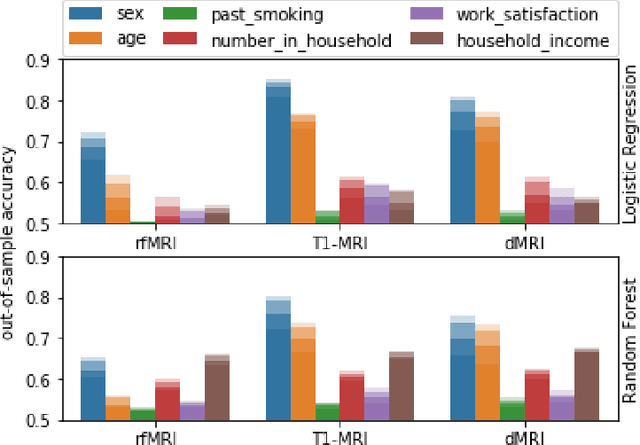
Abstract:High-quality data accumulation is now becoming ubiquitous in the health domain. There is increasing opportunity to exploit rich data from normal subjects to improve supervised estimators in specific diseases with notorious data scarcity. We demonstrate that low-dimensional embedding spaces can be derived from the UK Biobank population dataset and used to enhance data-scarce prediction of health indicators, lifestyle and demographic characteristics. Phenotype predictions facilitated by Variational Autoencoder manifolds typically scaled better with increasing unlabeled data than dimensionality reduction by PCA or Isomap. Performances gains from semisupervison approaches will probably become an important ingredient for various medical data science applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge