Marc-Andre Schulz
Brain-Semantoks: Learning Semantic Tokens of Brain Dynamics with a Self-Distilled Foundation Model
Dec 12, 2025Abstract:The development of foundation models for functional magnetic resonance imaging (fMRI) time series holds significant promise for predicting phenotypes related to disease and cognition. Current models, however, are often trained using a mask-and-reconstruct objective on small brain regions. This focus on low-level information leads to representations that are sensitive to noise and temporal fluctuations, necessitating extensive fine-tuning for downstream tasks. We introduce Brain-Semantoks, a self-supervised framework designed specifically to learn abstract representations of brain dynamics. Its architecture is built on two core innovations: a semantic tokenizer that aggregates noisy regional signals into robust tokens representing functional networks, and a self-distillation objective that enforces representational stability across time. We show that this objective is stabilized through a novel training curriculum, ensuring the model robustly learns meaningful features from low signal-to-noise time series. We demonstrate that learned representations enable strong performance on a variety of downstream tasks even when only using a linear probe. Furthermore, we provide comprehensive scaling analyses indicating more unlabeled data reliably results in out-of-distribution performance gains without domain adaptation.
Label scarcity in biomedicine: Data-rich latent factor discovery enhances phenotype prediction
Oct 12, 2021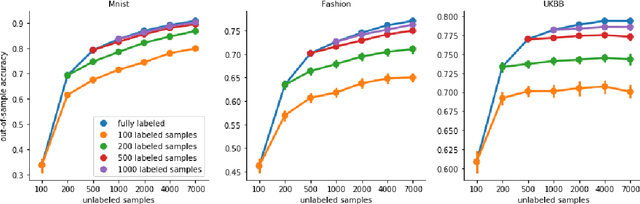
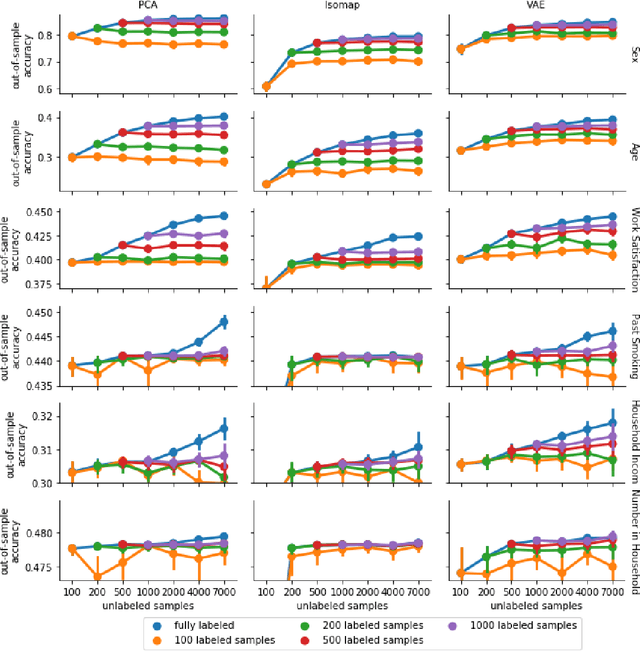
Abstract:High-quality data accumulation is now becoming ubiquitous in the health domain. There is increasing opportunity to exploit rich data from normal subjects to improve supervised estimators in specific diseases with notorious data scarcity. We demonstrate that low-dimensional embedding spaces can be derived from the UK Biobank population dataset and used to enhance data-scarce prediction of health indicators, lifestyle and demographic characteristics. Phenotype predictions facilitated by Variational Autoencoder manifolds typically scaled better with increasing unlabeled data than dimensionality reduction by PCA or Isomap. Performances gains from semisupervison approaches will probably become an important ingredient for various medical data science applications.
Clusters in Explanation Space: Inferring disease subtypes from model explanations
Dec 18, 2019
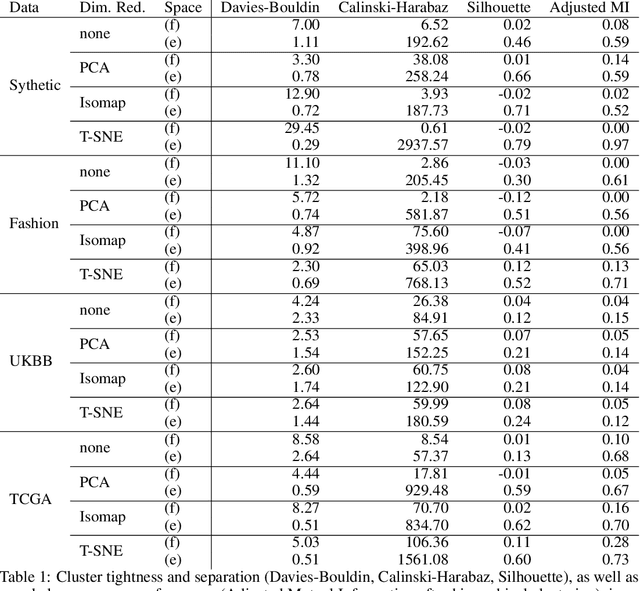
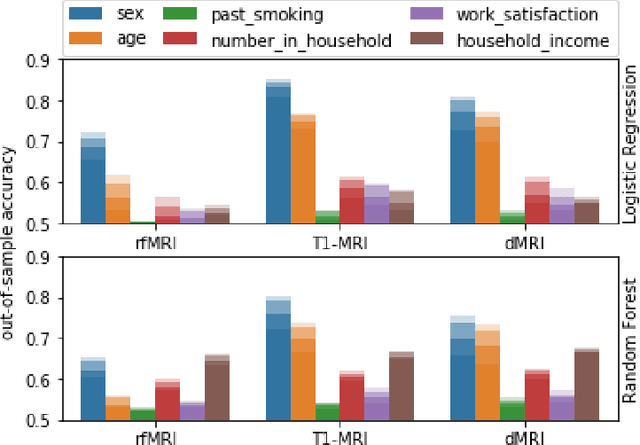
Abstract:Identification of disease subtypes and corresponding biomarkers can substantially improve clinical diagnosis and treatment selection. Discovering these subtypes in noisy, high dimensional biomedical data is often impossible for humans and challenging for machines. We introduce a new approach to facilitate the discovery of disease subtypes: Instead of analyzing the original data, we train a diagnostic classifier (healthy vs. diseased) and extract instance-wise explanations for the classifier's decisions. The distribution of instances in the explanation space of our diagnostic classifier amplifies the different reasons for belonging to the same class - resulting in a representation that is uniquely useful for discovering latent subtypes. We compare our ability to recover subtypes via cluster analysis on model explanations to classical cluster analysis on the original data. In multiple datasets with known ground-truth subclasses, most compellingly on UK Biobank brain imaging data and transcriptome data from the Cancer Genome Atlas, we show that cluster analysis on model explanations substantially outperforms the classical approach. While we believe clustering in explanation space to be particularly valuable for inferring disease subtypes, the method is more general and applicable to any kind of sub-type identification.
EMAP: Explanation by Minimal Adversarial Perturbation
Dec 02, 2019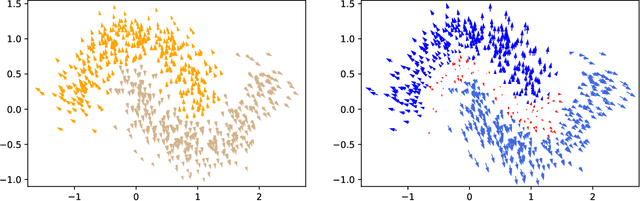
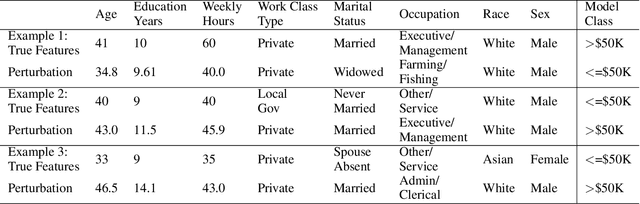
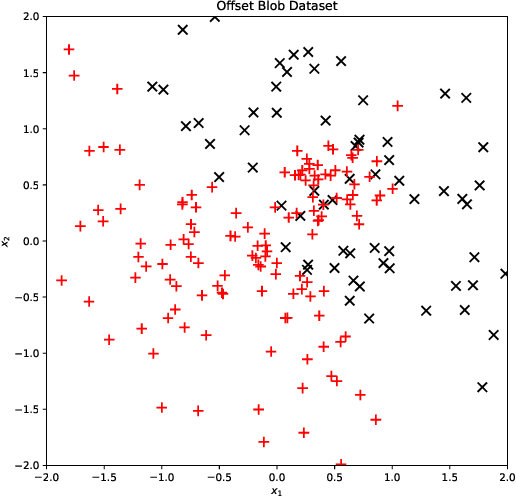
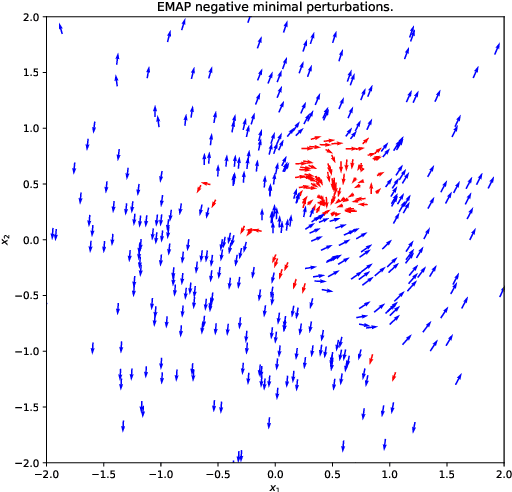
Abstract:Modern instance-based model-agnostic explanation methods (LIME, SHAP, L2X) are of great use in data-heavy industries for model diagnostics, and for end-user explanations. These methods generally return either a weighting or subset of input features as an explanation of the classification of an instance. An alternative literature argues instead that counterfactual instances provide a more useable characterisation of a black box classifier's decisions. We present EMAP, a neural network based approach which returns as Explanation the Minimal Adversarial Perturbation to an instance required to cause the underlying black box model to missclassify. We show that this approach combines the two paradigms, recovering the output of feature-weighting methods in continuous feature spaces, whilst also indicating the direction in which the nearest counterfactuals can be found. Our method also provides an implicit confidence estimate in its own explanations, adding a clarity to model diagnostics other methods lack. Additionally, EMAP improves upon the speed of sampling-based methods such as LIME by an order of magnitude, allowing for model explanations in time-critical applications, or at the dataset level, where sampling-based methods are infeasible. We extend our approach to categorical features using a partitioned Gumbel layer, and demonstrate its efficacy on several standard datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge