"cancer detection": models, code, and papers
Using Generative Models for Pediatric wbMRI
Jun 01, 2020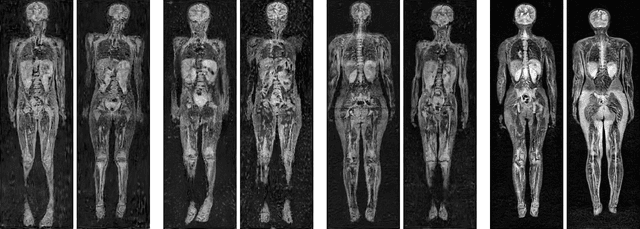
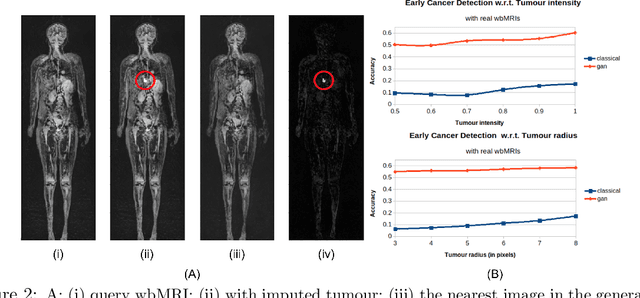
Early detection of cancer is key to a good prognosis and requires frequent testing, especially in pediatrics. Whole-body magnetic resonance imaging (wbMRI) is an essential part of several well-established screening protocols, with screening starting in early childhood. To date, machine learning (ML) has been used on wbMRI images to stage adult cancer patients. It is not possible to use such tools in pediatrics due to the changing bone signal throughout growth, the difficulty of obtaining these images in young children due to movement and limited compliance, and the rarity of positive cases. We evaluate the quality of wbMRI images generated using generative adversarial networks (GANs) trained on wbMRI data from The Hospital for Sick Children in Toronto. We use the Frchet Inception Distance (FID) metric, Domain Frchet Distance (DFD), and blind tests with a radiology fellow for evaluation. We demonstrate that StyleGAN2 provides the best performance in generating wbMRI images with respect to all three metrics.
Automatic Generation of Interpretable Lung Cancer Scoring Models from Chest X-Ray Images
Dec 10, 2020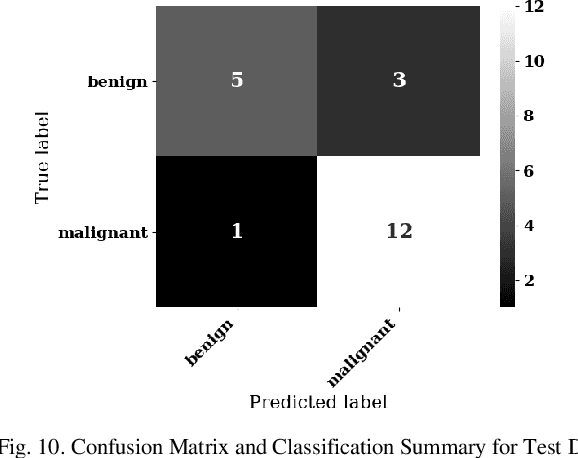
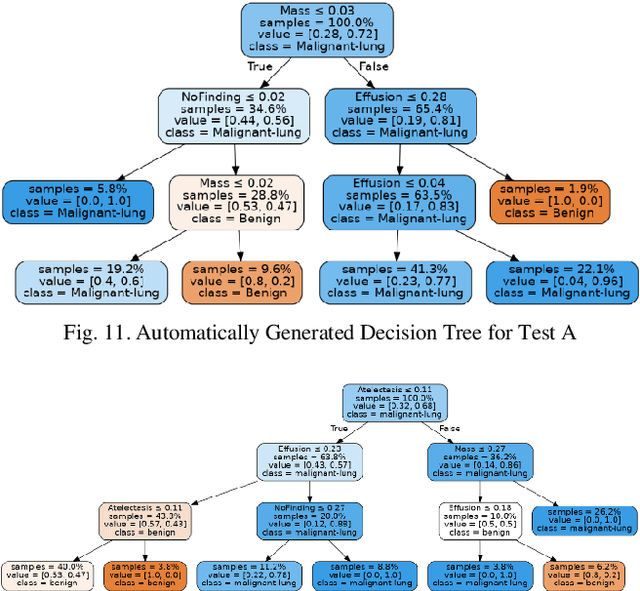
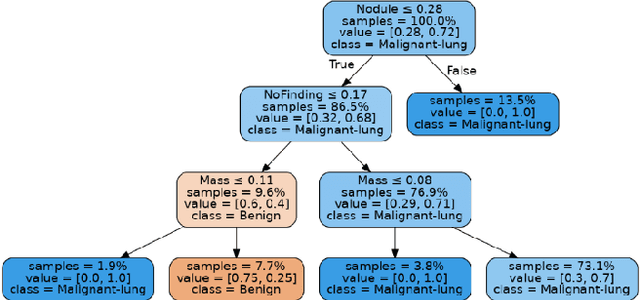
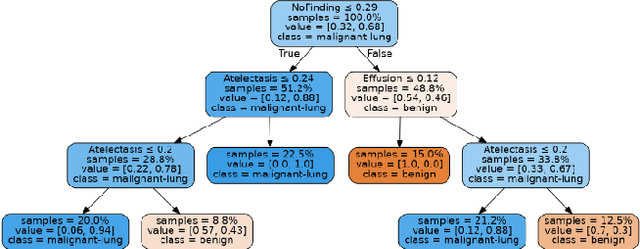
Lung cancer is the leading cause of cancer death and morbidity worldwide with early detection being the key to a positive patient prognosis. Although a multitude of studies have demonstrated that machine learning, and particularly deep learning, techniques are effective at automatically diagnosing lung cancer, these techniques have yet to be clinically approved and accepted/adopted by the medical community. Rather than attempting to provide an artificial 'second reading' we instead focus on the automatic creation of viable decision tree models from publicly available data using computer vision and machine learning techniques. For a small inferencing dataset, this method achieves a best accuracy over 84% with a positive predictive value of 83% for the malignant class. Furthermore, the decision trees created by this process may be considered as a starting point for refinement by medical experts into clinically usable multi-variate lung cancer scoring and diagnostic models.
Performance Comparison of Balanced and Unbalanced Cancer Datasets using Pre-Trained Convolutional Neural Network
Dec 10, 2020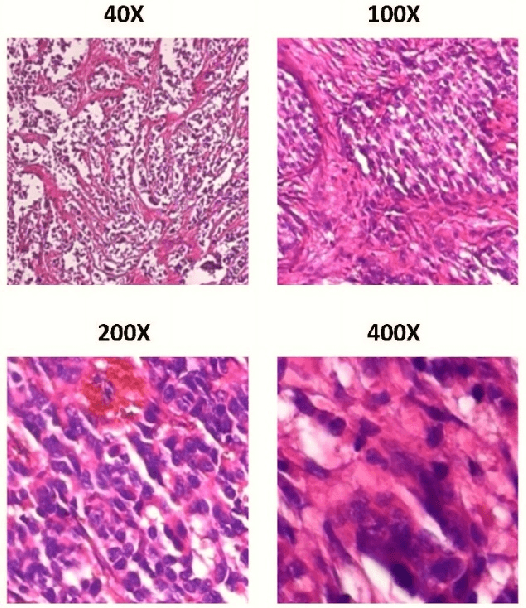
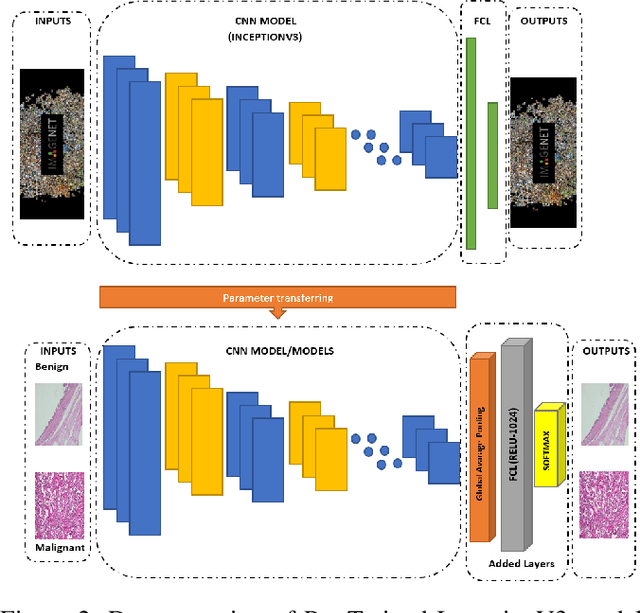
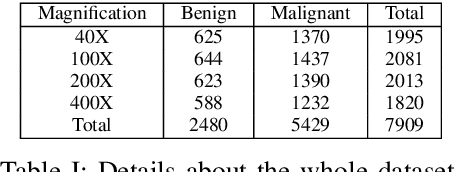
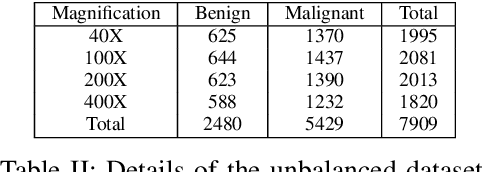
Cancer disease is one of the leading causes of death all over the world. Breast cancer, which is a common cancer disease especially in women, is quite common. The most important tool used for early detection of this cancer type, which requires a long process to establish a definitive diagnosis, is histopathological images taken by biopsy. These obtained images are examined by pathologists and a definitive diagnosis is made. It is quite common to detect this process with the help of a computer. Detection of benign or malignant tumors, especially by using data with different magnification rates, takes place in the literature. In this study, two different balanced and unbalanced study groups have been formed by using the histopathological data in the BreakHis data set. We have examined how the performances of balanced and unbalanced data sets change in detecting tumor type. In conclusion, in the study performed using the InceptionV3 convolution neural network model, 93.55% accuracy, 99.19% recall and 87.10% specificity values have been obtained for balanced data, while 89.75% accuracy, 82.89% recall and 91.51% specificity values have been obtained for unbalanced data. According to the results obtained in two different studies, the balance of the data increases the overall performance as well as the detection performance of both benign and malignant tumors. It can be said that the model trained with the help of data sets created in a balanced way will give pathology specialists higher and accurate results.
Mitosis Detection Under Limited Annotation: A Joint Learning Approach
Jun 17, 2020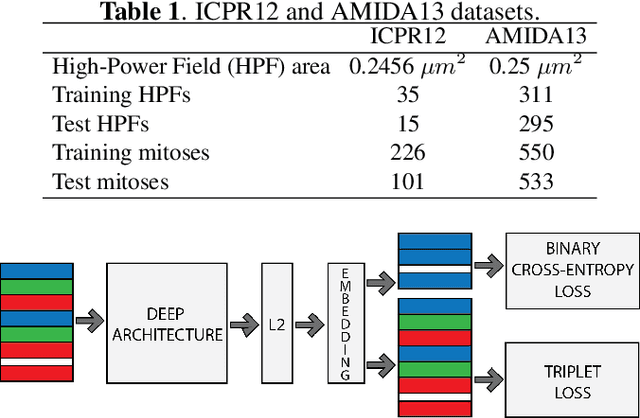
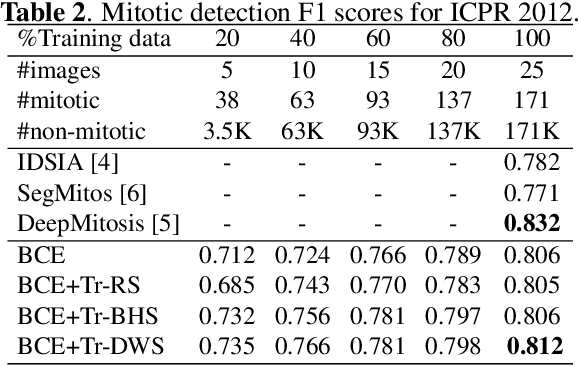
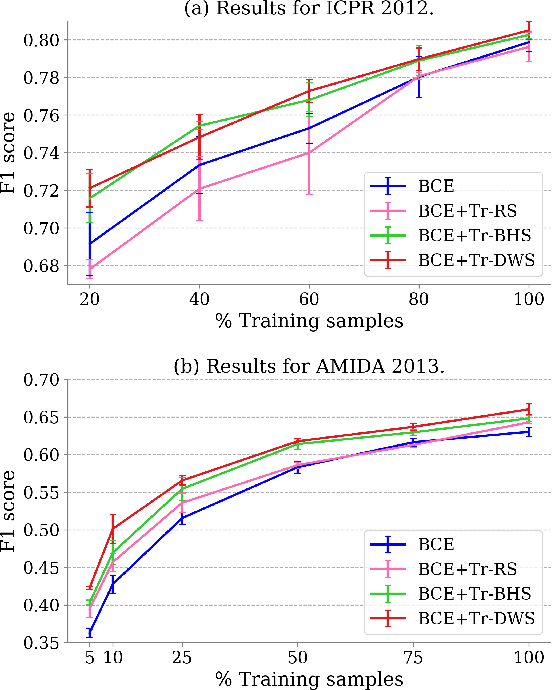
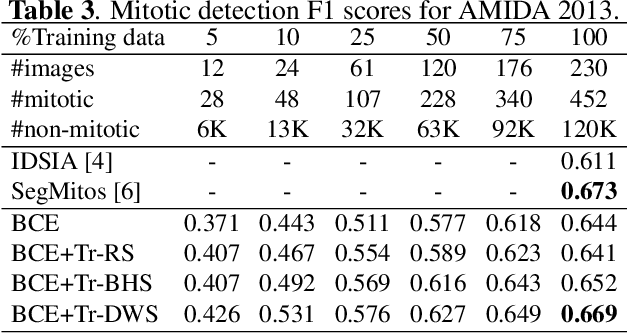
Mitotic counting is a vital prognostic marker of tumor proliferation in breast cancer. Deep learning-based mitotic detection is on par with pathologists, but it requires large labeled data for training. We propose a deep classification framework for enhancing mitosis detection by leveraging class label information, via softmax loss, and spatial distribution information among samples, via distance metric learning. We also investigate strategies towards steadily providing informative samples to boost the learning. The efficacy of the proposed framework is established through evaluation on ICPR 2012 and AMIDA 2013 mitotic data. Our framework significantly improves the detection with small training data and achieves on par or superior performance compared to state-of-the-art methods for using the entire training data.
Accurate Cell Segmentation in Digital Pathology Images via Attention Enforced Networks
Dec 27, 2020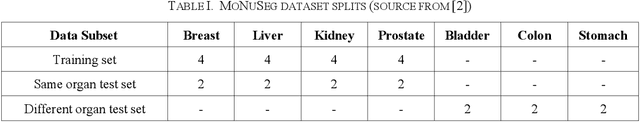
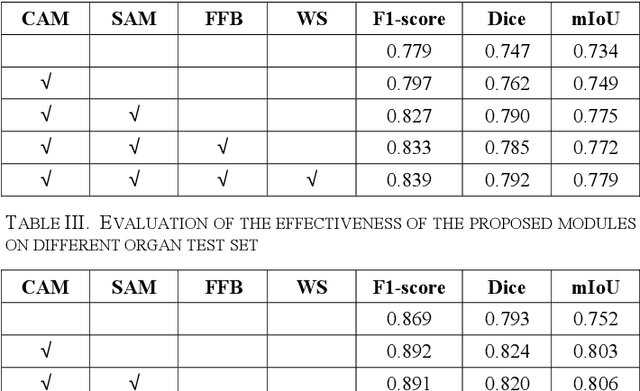
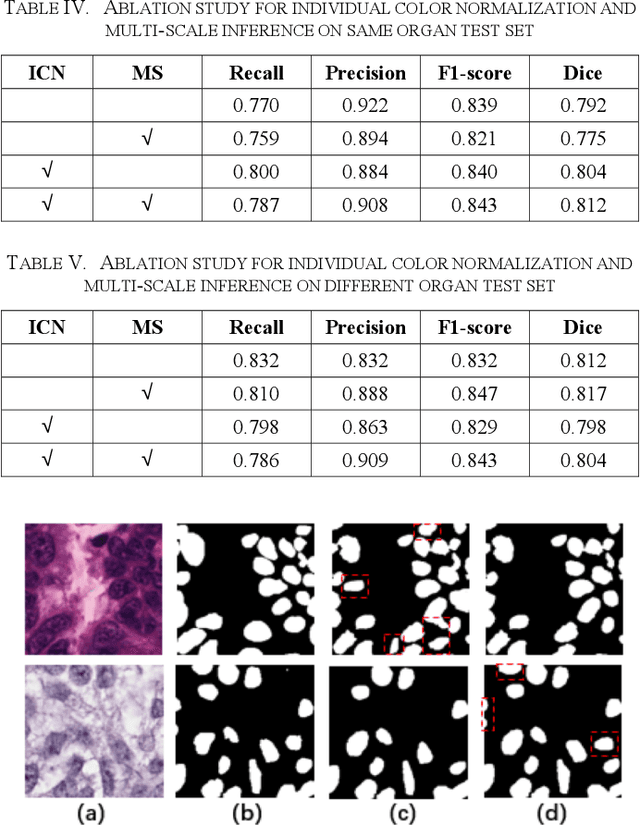
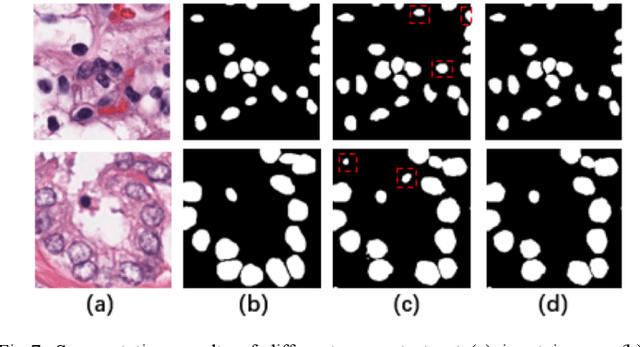
Automatic cell segmentation is an essential step in the pipeline of computer-aided diagnosis (CAD), such as the detection and grading of breast cancer. Accurate segmentation of cells can not only assist the pathologists to make a more precise diagnosis, but also save much time and labor. However, this task suffers from stain variation, cell inhomogeneous intensities, background clutters and cells from different tissues. To address these issues, we propose an Attention Enforced Network (AENet), which is built on spatial attention module and channel attention module, to integrate local features with global dependencies and weight effective channels adaptively. Besides, we introduce a feature fusion branch to bridge high-level and low-level features. Finally, the marker controlled watershed algorithm is applied to post-process the predicted segmentation maps for reducing the fragmented regions. In the test stage, we present an individual color normalization method to deal with the stain variation problem. We evaluate this model on the MoNuSeg dataset. The quantitative comparisons against several prior methods demonstrate the superiority of our approach.
A new semi-supervised self-training method for lung cancer prediction
Dec 17, 2020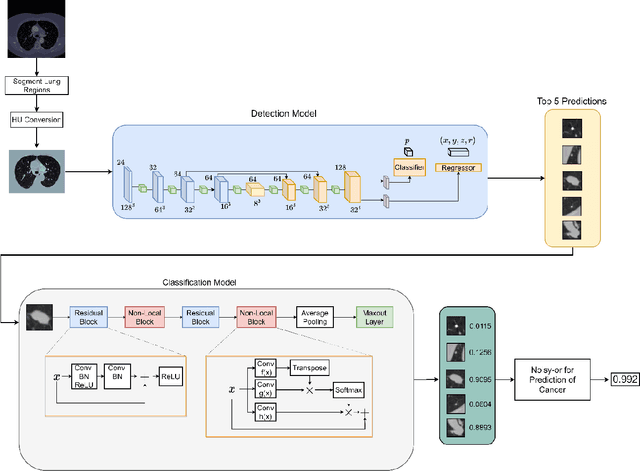
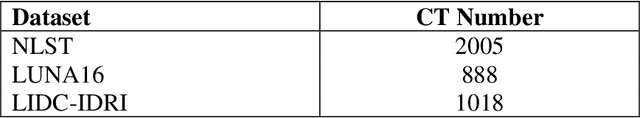
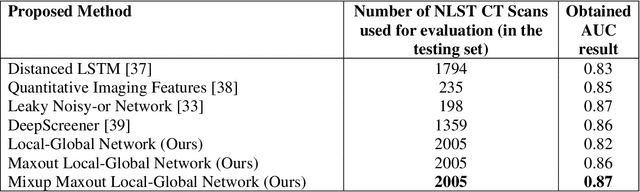
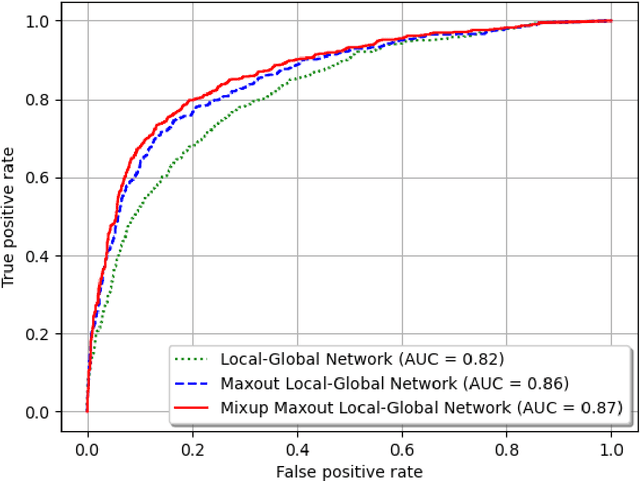
Background and Objective: Early detection of lung cancer is crucial as it has high mortality rate with patients commonly present with the disease at stage 3 and above. There are only relatively few methods that simultaneously detect and classify nodules from computed tomography (CT) scans. Furthermore, very few studies have used semi-supervised learning for lung cancer prediction. This study presents a complete end-to-end scheme to detect and classify lung nodules using the state-of-the-art Self-training with Noisy Student method on a comprehensive CT lung screening dataset of around 4,000 CT scans. Methods: We used three datasets, namely LUNA16, LIDC and NLST, for this study. We first utilise a three-dimensional deep convolutional neural network model to detect lung nodules in the detection stage. The classification model known as Maxout Local-Global Network uses non-local networks to detect global features including shape features, residual blocks to detect local features including nodule texture, and a Maxout layer to detect nodule variations. We trained the first Self-training with Noisy Student model to predict lung cancer on the unlabelled NLST datasets. Then, we performed Mixup regularization to enhance our scheme and provide robustness to erroneous labels. Results and Conclusions: Our new Mixup Maxout Local-Global network achieves an AUC of 0.87 on 2,005 completely independent testing scans from the NLST dataset. Our new scheme significantly outperformed the next highest performing method at the 5% significance level using DeLong's test (p = 0.0001). This study presents a new complete end-to-end scheme to predict lung cancer using Self-training with Noisy Student combined with Mixup regularization. On a completely independent dataset of 2,005 scans, we achieved state-of-the-art performance even with more images as compared to other methods.
Deep Learning for fully automatic detection, segmentation, and Gleason Grade estimation of prostate cancer in multiparametric Magnetic Resonance Images
Mar 24, 2021
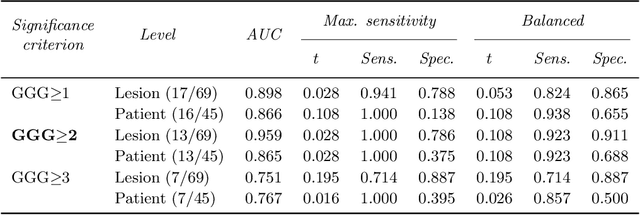
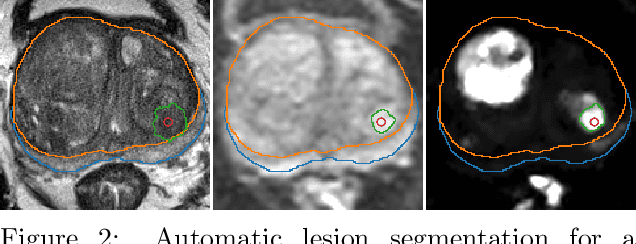
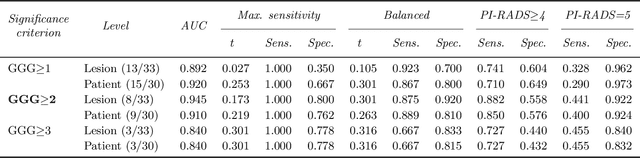
The emergence of multi-parametric magnetic resonance imaging (mpMRI) has had a profound impact on the diagnosis of prostate cancers (PCa), which is the most prevalent malignancy in males in the western world, enabling a better selection of patients for confirmation biopsy. However, analyzing these images is complex even for experts, hence opening an opportunity for computer-aided diagnosis systems to seize. This paper proposes a fully automatic system based on Deep Learning that takes a prostate mpMRI from a PCa-suspect patient and, by leveraging the Retina U-Net detection framework, locates PCa lesions, segments them, and predicts their most likely Gleason grade group (GGG). It uses 490 mpMRIs for training/validation, and 75 patients for testing from two different datasets: ProstateX and IVO (Valencia Oncology Institute Foundation). In the test set, it achieves an excellent lesion-level AUC/sensitivity/specificity for the GGG$\geq$2 significance criterion of 0.96/1.00/0.79 for the ProstateX dataset, and 0.95/1.00/0.80 for the IVO dataset. Evaluated at a patient level, the results are 0.87/1.00/0.375 in ProstateX, and 0.91/1.00/0.762 in IVO. Furthermore, on the online ProstateX grand challenge, the model obtained an AUC of 0.85 (0.87 when trained only on the ProstateX data, tying up with the original winner of the challenge). For expert comparison, IVO radiologist's PI-RADS 4 sensitivity/specificity were 0.88/0.56 at a lesion level, and 0.85/0.58 at a patient level. Additional subsystems for automatic prostate zonal segmentation and mpMRI non-rigid sequence registration were also employed to produce the final fully automated system. The code for the ProstateX-trained system has been made openly available at https://github.com/OscarPellicer/prostate_lesion_detection. We hope that this will represent a landmark for future research to use, compare and improve upon.
Application of S-Transform on Hyper kurtosis based Modified Duo Histogram Equalized DIC images for Pre-cancer Detection
Apr 30, 2015Our proposed hyper kurtosis based histogram equalized DIC images enhances the contrast by preserving the brightness. The evolution and development of precancerous activity among tissues are studied through S-transform (ST). The significant variations of amplitude spectra can be observed due to increased medium roughness from normal tissue were observed in time-frequency domain. The randomness and inhomogeneity of the tissue structures among human normal and different grades of DIC tissues is recognized by ST based timefrequency analysis. This study offers a simpler and better way to recognize the substantial changes among different stages of DIC tissues, which are reflected by spatial information containing within the inhomogeneity structures of different types of tissue.
Medico Multimedia Task at MediaEval 2020: Automatic Polyp Segmentation
Dec 30, 2020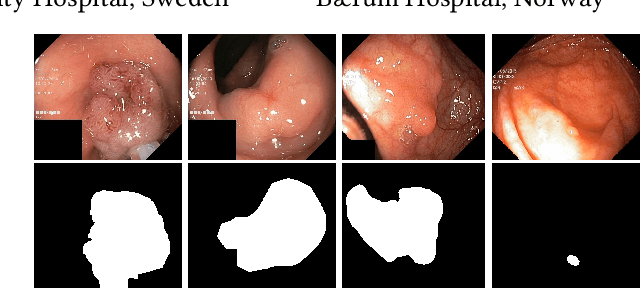

Colorectal cancer is the third most common cause of cancer worldwide. According to Global cancer statistics 2018, the incidence of colorectal cancer is increasing in both developing and developed countries. Early detection of colon anomalies such as polyps is important for cancer prevention, and automatic polyp segmentation can play a crucial role for this. Regardless of the recent advancement in early detection and treatment options, the estimated polyp miss rate is still around 20\%. Support via an automated computer-aided diagnosis system could be one of the potential solutions for the overlooked polyps. Such detection systems can help low-cost design solutions and save doctors time, which they could for example use to perform more patient examinations. In this paper, we introduce the 2020 Medico challenge, provide some information on related work and the dataset, describe the task and evaluation metrics, and discuss the necessity of organizing the Medico challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge