cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Attention-Enhanced Deep Learning Ensemble for Breast Density Classification in Mammography
Jul 08, 2025Breast density assessment is a crucial component of mammographic interpretation, with high breast density (BI-RADS categories C and D) representing both a significant risk factor for developing breast cancer and a technical challenge for tumor detection. This study proposes an automated deep learning system for robust binary classification of breast density (low: A/B vs. high: C/D) using the VinDr-Mammo dataset. We implemented and compared four advanced convolutional neural networks: ResNet18, ResNet50, EfficientNet-B0, and DenseNet121, each enhanced with channel attention mechanisms. To address the inherent class imbalance, we developed a novel Combined Focal Label Smoothing Loss function that integrates focal loss, label smoothing, and class-balanced weighting. Our preprocessing pipeline incorporated advanced techniques, including contrast-limited adaptive histogram equalization (CLAHE) and comprehensive data augmentation. The individual models were combined through an optimized ensemble voting approach, achieving superior performance (AUC: 0.963, F1-score: 0.952) compared to any single model. This system demonstrates significant potential to standardize density assessments in clinical practice, potentially improving screening efficiency and early cancer detection rates while reducing inter-observer variability among radiologists.
Towards Comprehensive Cellular Characterisation of H&E slides
Aug 13, 2025Cell detection, segmentation and classification are essential for analyzing tumor microenvironments (TME) on hematoxylin and eosin (H&E) slides. Existing methods suffer from poor performance on understudied cell types (rare or not present in public datasets) and limited cross-domain generalization. To address these shortcomings, we introduce HistoPLUS, a state-of-the-art model for cell analysis, trained on a novel curated pan-cancer dataset of 108,722 nuclei covering 13 cell types. In external validation across 4 independent cohorts, HistoPLUS outperforms current state-of-the-art models in detection quality by 5.2% and overall F1 classification score by 23.7%, while using 5x fewer parameters. Notably, HistoPLUS unlocks the study of 7 understudied cell types and brings significant improvements on 8 of 13 cell types. Moreover, we show that HistoPLUS robustly transfers to two oncology indications unseen during training. To support broader TME biomarker research, we release the model weights and inference code at https://github.com/owkin/histoplus/.
Cyst-X: AI-Powered Pancreatic Cancer Risk Prediction from Multicenter MRI in Centralized and Federated Learning
Jul 29, 2025



Pancreatic cancer is projected to become the second-deadliest malignancy in Western countries by 2030, highlighting the urgent need for better early detection. Intraductal papillary mucinous neoplasms (IPMNs), key precursors to pancreatic cancer, are challenging to assess with current guidelines, often leading to unnecessary surgeries or missed malignancies. We present Cyst-X, an AI framework that predicts IPMN malignancy using multicenter MRI data, leveraging MRI's superior soft tissue contrast over CT. Trained on 723 T1- and 738 T2-weighted scans from 764 patients across seven institutions, our models (AUC=0.82) significantly outperform both Kyoto guidelines (AUC=0.75) and expert radiologists. The AI-derived imaging features align with known clinical markers and offer biologically meaningful insights. We also demonstrate strong performance in a federated learning setting, enabling collaborative training without sharing patient data. To promote privacy-preserving AI development and improve IPMN risk stratification, the Cyst-X dataset is released as the first large-scale, multi-center pancreatic cysts MRI dataset.
UGPL: Uncertainty-Guided Progressive Learning for Evidence-Based Classification in Computed Tomography
Jul 18, 2025Accurate classification of computed tomography (CT) images is essential for diagnosis and treatment planning, but existing methods often struggle with the subtle and spatially diverse nature of pathological features. Current approaches typically process images uniformly, limiting their ability to detect localized abnormalities that require focused analysis. We introduce UGPL, an uncertainty-guided progressive learning framework that performs a global-to-local analysis by first identifying regions of diagnostic ambiguity and then conducting detailed examination of these critical areas. Our approach employs evidential deep learning to quantify predictive uncertainty, guiding the extraction of informative patches through a non-maximum suppression mechanism that maintains spatial diversity. This progressive refinement strategy, combined with an adaptive fusion mechanism, enables UGPL to integrate both contextual information and fine-grained details. Experiments across three CT datasets demonstrate that UGPL consistently outperforms state-of-the-art methods, achieving improvements of 3.29%, 2.46%, and 8.08% in accuracy for kidney abnormality, lung cancer, and COVID-19 detection, respectively. Our analysis shows that the uncertainty-guided component provides substantial benefits, with performance dramatically increasing when the full progressive learning pipeline is implemented. Our code is available at: https://github.com/shravan-18/UGPL
Reconsidering Explicit Longitudinal Mammography Alignment for Enhanced Breast Cancer Risk Prediction
Jun 24, 2025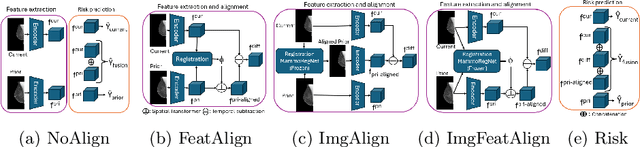
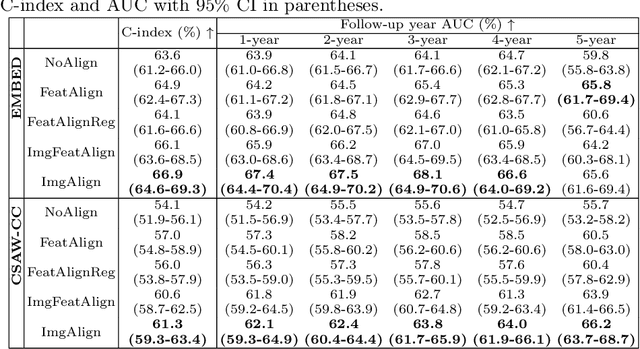
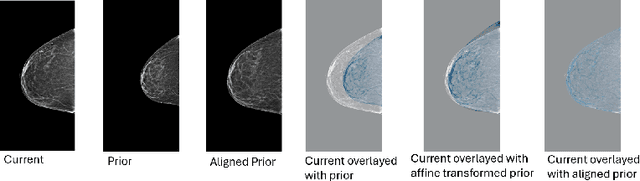
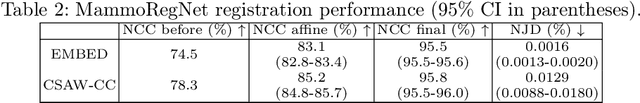
Regular mammography screening is essential for early breast cancer detection. Deep learning-based risk prediction methods have sparked interest to adjust screening intervals for high-risk groups. While early methods focused only on current mammograms, recent approaches leverage the temporal aspect of screenings to track breast tissue changes over time, requiring spatial alignment across different time points. Two main strategies for this have emerged: explicit feature alignment through deformable registration and implicit learned alignment using techniques like transformers, with the former providing more control. However, the optimal approach for explicit alignment in mammography remains underexplored. In this study, we provide insights into where explicit alignment should occur (input space vs. representation space) and if alignment and risk prediction should be jointly optimized. We demonstrate that jointly learning explicit alignment in representation space while optimizing risk estimation performance, as done in the current state-of-the-art approach, results in a trade-off between alignment quality and predictive performance and show that image-level alignment is superior to representation-level alignment, leading to better deformation field quality and enhanced risk prediction accuracy. The code is available at https://github.com/sot176/Longitudinal_Mammogram_Alignment.git.
Detection of Breast Cancer Lumpectomy Margin with SAM-incorporated Forward-Forward Contrastive Learning
Jun 26, 2025Complete removal of cancer tumors with a negative specimen margin during lumpectomy is essential in reducing breast cancer recurrence. However, 2D specimen radiography (SR), the current method used to assess intraoperative specimen margin status, has limited accuracy, resulting in nearly a quarter of patients requiring additional surgery. To address this, we propose a novel deep learning framework combining the Segment Anything Model (SAM) with Forward-Forward Contrastive Learning (FFCL), a pre-training strategy leveraging both local and global contrastive learning for patch-level classification of SR images. After annotating SR images with regions of known maligancy, non-malignant tissue, and pathology-confirmed margins, we pre-train a ResNet-18 backbone with FFCL to classify margin status, then reconstruct coarse binary masks to prompt SAM for refined tumor margin segmentation. Our approach achieved an AUC of 0.8455 for margin classification and segmented margins with a 27.4% improvement in Dice similarity over baseline models, while reducing inference time to 47 milliseconds per image. These results demonstrate that FFCL-SAM significantly enhances both the speed and accuracy of intraoperative margin assessment, with strong potential to reduce re-excision rates and improve surgical outcomes in breast cancer treatment. Our code is available at https://github.com/tbwa233/FFCL-SAM/.
MEGANet-W: A Wavelet-Driven Edge-Guided Attention Framework for Weak Boundary Polyp Detection
Jul 03, 2025Colorectal polyp segmentation is critical for early detection of colorectal cancer, yet weak and low contrast boundaries significantly limit automated accuracy. Existing deep models either blur fine edge details or rely on handcrafted filters that perform poorly under variable imaging conditions. We propose MEGANet-W, a Wavelet Driven Edge Guided Attention Network that injects directional, parameter free Haar wavelet edge maps into each decoder stage to recalibrate semantic features. Our two main contributions are: (1) a two-level Haar wavelet head for multi orientation edge extraction; and (2) Wavelet Edge Guided Attention (WEGA) modules that fuse wavelet cues with reverse and input branches. On five public polyp datasets, MEGANetW consistently outperforms existing methods, improving mIoU by up to 2.3% and mDice by 1.2%, while introducing no additional learnable parameters.
EndoFinder: Online Lesion Retrieval for Explainable Colorectal Polyp Diagnosis Leveraging Latent Scene Representations
Jul 23, 2025Colorectal cancer (CRC) remains a leading cause of cancer-related mortality, underscoring the importance of timely polyp detection and diagnosis. While deep learning models have improved optical-assisted diagnostics, they often demand extensive labeled datasets and yield "black-box" outputs with limited interpretability. In this paper, we propose EndoFinder, an online polyp retrieval framework that leverages multi-view scene representations for explainable and scalable CRC diagnosis. First, we develop a Polyp-aware Image Encoder by combining contrastive learning and a reconstruction task, guided by polyp segmentation masks. This self-supervised approach captures robust features without relying on large-scale annotated data. Next, we treat each polyp as a three-dimensional "scene" and introduce a Scene Representation Transformer, which fuses multiple views of the polyp into a single latent representation. By discretizing this representation through a hashing layer, EndoFinder enables real-time retrieval from a compiled database of historical polyp cases, where diagnostic information serves as interpretable references for new queries. We evaluate EndoFinder on both public and newly collected polyp datasets for re-identification and pathology classification. Results show that EndoFinder outperforms existing methods in accuracy while providing transparent, retrieval-based insights for clinical decision-making. By contributing a novel dataset and a scalable, explainable framework, our work addresses key challenges in polyp diagnosis and offers a promising direction for more efficient AI-driven colonoscopy workflows. The source code is available at https://github.com/ku262/EndoFinder-Scene.
Detecting malignant dynamics on very few blood sample using signature coefficients
Jun 10, 2025Recent discoveries have suggested that the promising avenue of using circulating tumor DNA (ctDNA) levels in blood samples provides reasonable accuracy for cancer monitoring, with extremely low burden on the patient's side. It is known that the presence of ctDNA can result from various mechanisms leading to DNA release from cells, such as apoptosis, necrosis or active secretion. One key idea in recent cancer monitoring studies is that monitoring the dynamics of ctDNA levels might be sufficient for early multi-cancer detection. This interesting idea has been turned into commercial products, e.g. in the company named GRAIL. In the present work, we propose to explore the use of Signature theory for detecting aggressive cancer tumors based on the analysis of blood samples. Our approach combines tools from continuous time Markov modelling for the dynamics of ctDNA levels in the blood, with Signature theory for building efficient testing procedures. Signature theory is a topic of growing interest in the Machine Learning community (see Chevyrev2016 and Fermanian2021), which is now recognised as a powerful feature extraction tool for irregularly sampled signals. The method proposed in the present paper is shown to correctly address the challenging problem of overcoming the inherent data scarsity due to the extremely small number of blood samples per patient. The relevance of our approach is illustrated with extensive numerical experiments that confirm the efficiency of the proposed pipeline.
Integrating Complexity and Biological Realism: High-Performance Spiking Neural Networks for Breast Cancer Detection
Jun 06, 2025Spiking Neural Networks (SNNs) event-driven nature enables efficient encoding of spatial and temporal features, making them suitable for dynamic time-dependent data processing. Despite their biological relevance, SNNs have seen limited application in medical image recognition due to difficulties in matching the performance of conventional deep learning models. To address this, we propose a novel breast cancer classification approach that combines SNNs with Lempel-Ziv Complexity (LZC) a computationally efficient measure of sequence complexity. LZC enhances the interpretability and accuracy of spike-based models by capturing structural patterns in neural activity. Our study explores both biophysical Leaky Integrate-and-Fire (LIF) and probabilistic Levy-Baxter (LB) neuron models under supervised, unsupervised, and hybrid learning regimes. Experiments were conducted on the Breast Cancer Wisconsin dataset using numerical features derived from medical imaging. LB-based models consistently exceeded 90.00% accuracy, while LIF-based models reached over 85.00%. The highest accuracy of 98.25% was achieved using an ANN-to-SNN conversion method applied to both neuron models comparable to traditional deep learning with back-propagation, but at up to 100 times lower computational cost. This hybrid approach merges deep learning performance with the efficiency and plausibility of SNNs, yielding top results at lower computational cost. We hypothesize that the synergy between temporal-coding, spike-sparsity, and LZC-driven complexity analysis enables more-efficient feature extraction. Our findings demonstrate that SNNs combined with LZC offer promising, biologically plausible alternative to conventional neural networks in medical diagnostics, particularly for resource-constrained or real-time systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge