Ali Braytee
Towards Automated Differential Diagnosis of Skin Diseases Using Deep Learning and Imbalance-Aware Strategies
Jan 01, 2026Abstract:As dermatological conditions become increasingly common and the availability of dermatologists remains limited, there is a growing need for intelligent tools to support both patients and clinicians in the timely and accurate diagnosis of skin diseases. In this project, we developed a deep learning based model for the classification and diagnosis of skin conditions. By leveraging pretraining on publicly available skin disease image datasets, our model effectively extracted visual features and accurately classified various dermatological cases. Throughout the project, we refined the model architecture, optimized data preprocessing workflows, and applied targeted data augmentation techniques to improve overall performance. The final model, based on the Swin Transformer, achieved a prediction accuracy of 87.71 percent across eight skin lesion classes on the ISIC2019 dataset. These results demonstrate the model's potential as a diagnostic support tool for clinicians and a self assessment aid for patients.
Benchmarking Preprocessing and Integration Methods in Single-Cell Genomics
Jan 01, 2026Abstract:Single-cell data analysis has the potential to revolutionize personalized medicine by characterizing disease-associated molecular changes at the single-cell level. Advanced single-cell multimodal assays can now simultaneously measure various molecules (e.g., DNA, RNA, Protein) across hundreds of thousands of individual cells, providing a comprehensive molecular readout. A significant analytical challenge is integrating single-cell measurements across different modalities. Various methods have been developed to address this challenge, but there has been no systematic evaluation of these techniques with different preprocessing strategies. This study examines a general pipeline for single-cell data analysis, which includes normalization, data integration, and dimensionality reduction. The performance of different algorithm combinations often depends on the dataset sizes and characteristics. We evaluate six datasets across diverse modalities, tissues, and organisms using three metrics: Silhouette Coefficient Score, Adjusted Rand Index, and Calinski-Harabasz Index. Our experiments involve combinations of seven normalization methods, four dimensional reduction methods, and five integration methods. The results show that Seurat and Harmony excel in data integration, with Harmony being more time-efficient, especially for large datasets. UMAP is the most compatible dimensionality reduction method with the integration techniques, and the choice of normalization method varies depending on the integration method used.
RSVLM-QA: A Benchmark Dataset for Remote Sensing Vision Language Model-based Question Answering
Aug 11, 2025Abstract:Visual Question Answering (VQA) in remote sensing (RS) is pivotal for interpreting Earth observation data. However, existing RS VQA datasets are constrained by limitations in annotation richness, question diversity, and the assessment of specific reasoning capabilities. This paper introduces RSVLM-QA dataset, a new large-scale, content-rich VQA dataset for the RS domain. RSVLM-QA is constructed by integrating data from several prominent RS segmentation and detection datasets: WHU, LoveDA, INRIA, and iSAID. We employ an innovative dual-track annotation generation pipeline. Firstly, we leverage Large Language Models (LLMs), specifically GPT-4.1, with meticulously designed prompts to automatically generate a suite of detailed annotations including image captions, spatial relations, and semantic tags, alongside complex caption-based VQA pairs. Secondly, to address the challenging task of object counting in RS imagery, we have developed a specialized automated process that extracts object counts directly from the original segmentation data; GPT-4.1 then formulates natural language answers from these counts, which are paired with preset question templates to create counting QA pairs. RSVLM-QA comprises 13,820 images and 162,373 VQA pairs, featuring extensive annotations and diverse question types. We provide a detailed statistical analysis of the dataset and a comparison with existing RS VQA benchmarks, highlighting the superior depth and breadth of RSVLM-QA's annotations. Furthermore, we conduct benchmark experiments on Six mainstream Vision Language Models (VLMs), demonstrating that RSVLM-QA effectively evaluates and challenges the understanding and reasoning abilities of current VLMs in the RS domain. We believe RSVLM-QA will serve as a pivotal resource for the RS VQA and VLM research communities, poised to catalyze advancements in the field.
Simplified Swarm Learning Framework for Robust and Scalable Diagnostic Services in Cancer Histopathology
Apr 23, 2025Abstract:The complexities of healthcare data, including privacy concerns, imbalanced datasets, and interoperability issues, necessitate innovative machine learning solutions. Swarm Learning (SL), a decentralized alternative to Federated Learning, offers privacy-preserving distributed training, but its reliance on blockchain technology hinders accessibility and scalability. This paper introduces a \textit{Simplified Peer-to-Peer Swarm Learning (P2P-SL) Framework} tailored for resource-constrained environments. By eliminating blockchain dependencies and adopting lightweight peer-to-peer communication, the proposed framework ensures robust model synchronization while maintaining data privacy. Applied to cancer histopathology, the framework integrates optimized pre-trained models, such as TorchXRayVision, enhanced with DenseNet decoders, to improve diagnostic accuracy. Extensive experiments demonstrate the framework's efficacy in handling imbalanced and biased datasets, achieving comparable performance to centralized models while preserving privacy. This study paves the way for democratizing advanced machine learning in healthcare, offering a scalable, accessible, and efficient solution for privacy-sensitive diagnostic applications.
Detecting and Understanding Hateful Contents in Memes Through Captioning and Visual Question-Answering
Apr 23, 2025Abstract:Memes are widely used for humor and cultural commentary, but they are increasingly exploited to spread hateful content. Due to their multimodal nature, hateful memes often evade traditional text-only or image-only detection systems, particularly when they employ subtle or coded references. To address these challenges, we propose a multimodal hate detection framework that integrates key components: OCR to extract embedded text, captioning to describe visual content neutrally, sub-label classification for granular categorization of hateful content, RAG for contextually relevant retrieval, and VQA for iterative analysis of symbolic and contextual cues. This enables the framework to uncover latent signals that simpler pipelines fail to detect. Experimental results on the Facebook Hateful Memes dataset reveal that the proposed framework exceeds the performance of unimodal and conventional multimodal models in both accuracy and AUC-ROC.
DualPrompt-MedCap: A Dual-Prompt Enhanced Approach for Medical Image Captioning
Apr 13, 2025



Abstract:Medical image captioning via vision-language models has shown promising potential for clinical diagnosis assistance. However, generating contextually relevant descriptions with accurate modality recognition remains challenging. We present DualPrompt-MedCap, a novel dual-prompt enhancement framework that augments Large Vision-Language Models (LVLMs) through two specialized components: (1) a modality-aware prompt derived from a semi-supervised classification model pretrained on medical question-answer pairs, and (2) a question-guided prompt leveraging biomedical language model embeddings. To address the lack of captioning ground truth, we also propose an evaluation framework that jointly considers spatial-semantic relevance and medical narrative quality. Experiments on multiple medical datasets demonstrate that DualPrompt-MedCap outperforms the baseline BLIP-3 by achieving a 22% improvement in modality recognition accuracy while generating more comprehensive and question-aligned descriptions. Our method enables the generation of clinically accurate reports that can serve as medical experts' prior knowledge and automatic annotations for downstream vision-language tasks.
AeroLite: Tag-Guided Lightweight Generation of Aerial Image Captions
Apr 13, 2025Abstract:Accurate and automated captioning of aerial imagery is crucial for applications like environmental monitoring, urban planning, and disaster management. However, this task remains challenging due to complex spatial semantics and domain variability. To address these issues, we introduce \textbf{AeroLite}, a lightweight, tag-guided captioning framework designed to equip small-scale language models (1--3B parameters) with robust and interpretable captioning capabilities specifically for remote sensing images. \textbf{AeroLite} leverages GPT-4o to generate a large-scale, semantically rich pseudo-caption dataset by integrating multiple remote sensing benchmarks, including DLRSD, iSAID, LoveDA, WHU, and RSSCN7. To explicitly capture key semantic elements such as orientation and land-use types, AeroLite employs natural language processing techniques to extract relevant semantic tags. These tags are then learned by a dedicated multi-label CLIP encoder, ensuring precise semantic predictions. To effectively fuse visual and semantic information, we propose a novel bridging multilayer perceptron (MLP) architecture, aligning semantic tags with visual embeddings while maintaining minimal computational overhead. AeroLite's flexible design also enables seamless integration with various pretrained large language models. We adopt a two-stage LoRA-based training approach: the initial stage leverages our pseudo-caption dataset to capture broad remote sensing semantics, followed by fine-tuning on smaller, curated datasets like UCM and Sydney Captions to refine domain-specific alignment. Experimental evaluations demonstrate that AeroLite surpasses significantly larger models (e.g., 13B parameters) in standard captioning metrics, including BLEU and METEOR, while maintaining substantially lower computational costs.
Vision Transformers with Autoencoders and Explainable AI for Cancer Patient Risk Stratification Using Whole Slide Imaging
Apr 08, 2025



Abstract:Cancer remains one of the leading causes of mortality worldwide, necessitating accurate diagnosis and prognosis. Whole Slide Imaging (WSI) has become an integral part of clinical workflows with advancements in digital pathology. While various studies have utilized WSIs, their extracted features may not fully capture the most relevant pathological information, and their lack of interpretability limits clinical adoption. In this paper, we propose PATH-X, a framework that integrates Vision Transformers (ViT) and Autoencoders with SHAP (Shapley Additive Explanations) to enhance model explainability for patient stratification and risk prediction using WSIs from The Cancer Genome Atlas (TCGA). A representative image slice is selected from each WSI, and numerical feature embeddings are extracted using Google's pre-trained ViT. These features are then compressed via an autoencoder and used for unsupervised clustering and classification tasks. Kaplan-Meier survival analysis is applied to evaluate stratification into two and three risk groups. SHAP is used to identify key contributing features, which are mapped onto histopathological slices to provide spatial context. PATH-X demonstrates strong performance in breast and glioma cancers, where a sufficient number of WSIs enabled robust stratification. However, performance in lung cancer was limited due to data availability, emphasizing the need for larger datasets to enhance model reliability and clinical applicability.
FedSAF: A Federated Learning Framework for Enhanced Gastric Cancer Detection and Privacy Preservation
Mar 20, 2025



Abstract:Gastric cancer is one of the most commonly diagnosed cancers and has a high mortality rate. Due to limited medical resources, developing machine learning models for gastric cancer recognition provides an efficient solution for medical institutions. However, such models typically require large sample sizes for training and testing, which can challenge patient privacy. Federated learning offers an effective alternative by enabling model training across multiple institutions without sharing sensitive patient data. This paper addresses the limited sample size of publicly available gastric cancer data with a modified data processing method. This paper introduces FedSAF, a novel federated learning algorithm designed to improve the performance of existing methods, particularly in non-independent and identically distributed (non-IID) data scenarios. FedSAF incorporates attention-based message passing and the Fisher Information Matrix to enhance model accuracy, while a model splitting function reduces computation and transmission costs. Hyperparameter tuning and ablation studies demonstrate the effectiveness of this new algorithm, showing improvements in test accuracy on gastric cancer datasets, with FedSAF outperforming existing federated learning methods like FedAMP, FedAvg, and FedProx. The framework's robustness and generalization ability were further validated across additional datasets (SEED, BOT, FashionMNIST, and CIFAR-10), achieving high performance in diverse environments.
Enhancing Sentiment Analysis through Multimodal Fusion: A BERT-DINOv2 Approach
Mar 11, 2025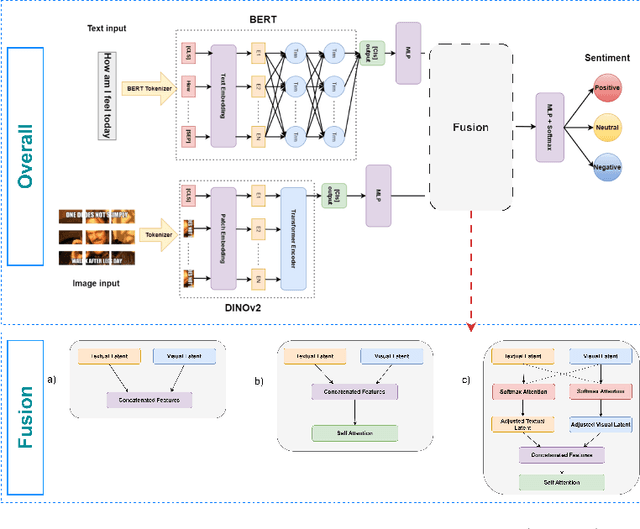
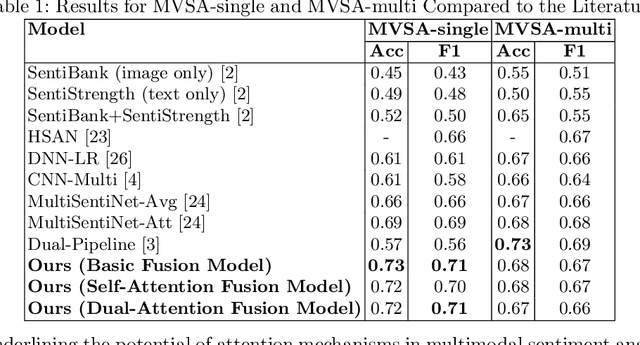
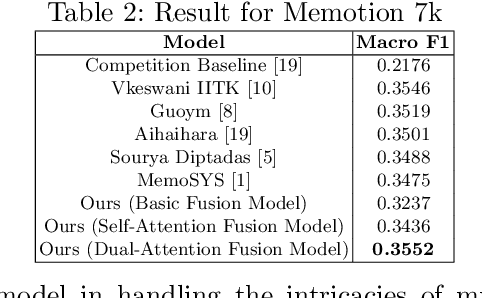
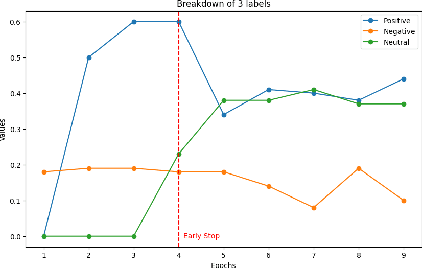
Abstract:Multimodal sentiment analysis enhances conventional sentiment analysis, which traditionally relies solely on text, by incorporating information from different modalities such as images, text, and audio. This paper proposes a novel multimodal sentiment analysis architecture that integrates text and image data to provide a more comprehensive understanding of sentiments. For text feature extraction, we utilize BERT, a natural language processing model. For image feature extraction, we employ DINOv2, a vision-transformer-based model. The textual and visual latent features are integrated using proposed fusion techniques, namely the Basic Fusion Model, Self Attention Fusion Model, and Dual Attention Fusion Model. Experiments on three datasets, Memotion 7k dataset, MVSA single dataset, and MVSA multi dataset, demonstrate the viability and practicality of the proposed multimodal architecture.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge