Plant Phenotyping
Papers and Code
Changepoint Detection As Model Selection: A General Framework
Jan 30, 2026This dissertation presents a general framework for changepoint detection based on L0 model selection. The core method, Iteratively Reweighted Fused Lasso (IRFL), improves upon the generalized lasso by adaptively reweighting penalties to enhance support recovery and minimize criteria such as the Bayesian Information Criterion (BIC). The approach allows for flexible modeling of seasonal patterns, linear and quadratic trends, and autoregressive dependence in the presence of changepoints. Simulation studies demonstrate that IRFL achieves accurate changepoint detection across a wide range of challenging scenarios, including those involving nuisance factors such as trends, seasonal patterns, and serially correlated errors. The framework is further extended to image data, where it enables edge-preserving denoising and segmentation, with applications spanning medical imaging and high-throughput plant phenotyping. Applications to real-world data demonstrate IRFL's utility. In particular, analysis of the Mauna Loa CO2 time series reveals changepoints that align with volcanic eruptions and ENSO events, yielding a more accurate trend decomposition than ordinary least squares. Overall, IRFL provides a robust, extensible tool for detecting structural change in complex data.
DepthCropSeg++: Scaling a Crop Segmentation Foundation Model With Depth-Labeled Data
Jan 18, 2026DepthCropSeg++: a foundation model for crop segmentation, capable of segmenting different crop species under open in-field environment. Crop segmentation is a fundamental task for modern agriculture, which closely relates to many downstream tasks such as plant phenotyping, density estimation, and weed control. In the era of foundation models, a number of generic large language and vision models have been developed. These models have demonstrated remarkable real world generalization due to significant model capacity and largescale datasets. However, current crop segmentation models mostly learn from limited data due to expensive pixel-level labelling cost, often performing well only under specific crop types or controlled environment. In this work, we follow the vein of our previous work DepthCropSeg, an almost unsupervised approach to crop segmentation, to scale up a cross-species and crossscene crop segmentation dataset, with 28,406 images across 30+ species and 15 environmental conditions. We also build upon a state-of-the-art semantic segmentation architecture ViT-Adapter architecture, enhance it with dynamic upsampling for improved detail awareness, and train the model with a two-stage selftraining pipeline. To systematically validate model performance, we conduct comprehensive experiments to justify the effectiveness and generalization capabilities across multiple crop datasets. Results demonstrate that DepthCropSeg++ achieves 93.11% mIoU on a comprehensive testing set, outperforming both supervised baselines and general-purpose vision foundation models like Segmentation Anything Model (SAM) by significant margins (+0.36% and +48.57% respectively). The model particularly excels in challenging scenarios including night-time environment (86.90% mIoU), high-density canopies (90.09% mIoU), and unseen crop varieties (90.09% mIoU), indicating a new state of the art for crop segmentation.
* 13 pages, 15 figures and 7 tables
LeafTrackNet: A Deep Learning Framework for Robust Leaf Tracking in Top-Down Plant Phenotyping
Dec 15, 2025High resolution phenotyping at the level of individual leaves offers fine-grained insights into plant development and stress responses. However, the full potential of accurate leaf tracking over time remains largely unexplored due to the absence of robust tracking methods-particularly for structurally complex crops such as canola. Existing plant-specific tracking methods are typically limited to small-scale species or rely on constrained imaging conditions. In contrast, generic multi-object tracking (MOT) methods are not designed for dynamic biological scenes. Progress in the development of accurate leaf tracking models has also been hindered by a lack of large-scale datasets captured under realistic conditions. In this work, we introduce CanolaTrack, a new benchmark dataset comprising 5,704 RGB images with 31,840 annotated leaf instances spanning the early growth stages of 184 canola plants. To enable accurate leaf tracking over time, we introduce LeafTrackNet, an efficient framework that combines a YOLOv10-based leaf detector with a MobileNetV3-based embedding network. During inference, leaf identities are maintained over time through an embedding-based memory association strategy. LeafTrackNet outperforms both plant-specific trackers and state-of-the-art MOT baselines, achieving a 9% HOTA improvement on CanolaTrack. With our work we provide a new standard for leaf-level tracking under realistic conditions and we provide CanolaTrack - the largest dataset for leaf tracking in agriculture crops, which will contribute to future research in plant phenotyping. Our code and dataset are publicly available at https://github.com/shl-shawn/LeafTrackNet.
GaussianPlant: Structure-aligned Gaussian Splatting for 3D Reconstruction of Plants
Dec 16, 2025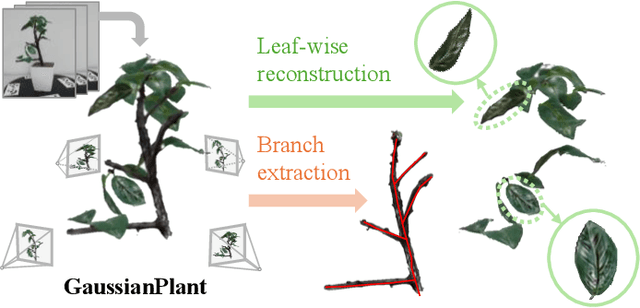

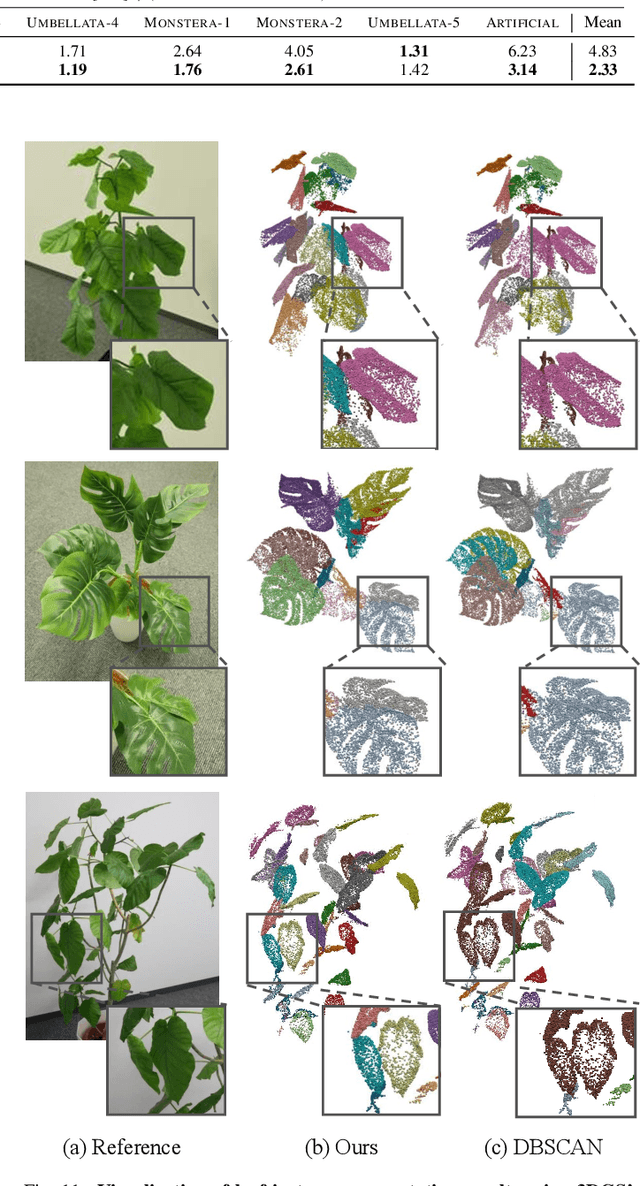

We present a method for jointly recovering the appearance and internal structure of botanical plants from multi-view images based on 3D Gaussian Splatting (3DGS). While 3DGS exhibits robust reconstruction of scene appearance for novel-view synthesis, it lacks structural representations underlying those appearances (e.g., branching patterns of plants), which limits its applicability to tasks such as plant phenotyping. To achieve both high-fidelity appearance and structural reconstruction, we introduce GaussianPlant, a hierarchical 3DGS representation, which disentangles structure and appearance. Specifically, we employ structure primitives (StPs) to explicitly represent branch and leaf geometry, and appearance primitives (ApPs) to the plants' appearance using 3D Gaussians. StPs represent a simplified structure of the plant, i.e., modeling branches as cylinders and leaves as disks. To accurately distinguish the branches and leaves, StP's attributes (i.e., branches or leaves) are optimized in a self-organized manner. ApPs are bound to each StP to represent the appearance of branches or leaves as in conventional 3DGS. StPs and ApPs are jointly optimized using a re-rendering loss on the input multi-view images, as well as the gradient flow from ApP to StP using the binding correspondence information. We conduct experiments to qualitatively evaluate the reconstruction accuracy of both appearance and structure, as well as real-world experiments to qualitatively validate the practical performance. Experiments show that the GaussianPlant achieves both high-fidelity appearance reconstruction via ApPs and accurate structural reconstruction via StPs, enabling the extraction of branch structure and leaf instances.
Generative diffusion models for agricultural AI: plant image generation, indoor-to-outdoor translation, and expert preference alignment
Dec 22, 2025



The success of agricultural artificial intelligence depends heavily on large, diverse, and high-quality plant image datasets, yet collecting such data in real field conditions is costly, labor intensive, and seasonally constrained. This paper investigates diffusion-based generative modeling to address these challenges through plant image synthesis, indoor-to-outdoor translation, and expert preference aligned fine tuning. First, a Stable Diffusion model is fine tuned on captioned indoor and outdoor plant imagery to generate realistic, text conditioned images of canola and soybean. Evaluation using Inception Score, Frechet Inception Distance, and downstream phenotype classification shows that synthetic images effectively augment training data and improve accuracy. Second, we bridge the gap between high resolution indoor datasets and limited outdoor imagery using DreamBooth-based text inversion and image guided diffusion, generating translated images that enhance weed detection and classification with YOLOv8. Finally, a preference guided fine tuning framework trains a reward model on expert scores and applies reward weighted updates to produce more stable and expert aligned outputs. Together, these components demonstrate a practical pathway toward data efficient generative pipelines for agricultural AI.
ST-DETrack: Identity-Preserving Branch Tracking in Entangled Plant Canopies via Dual Spatiotemporal Evidence
Dec 17, 2025



Automated extraction of individual plant branches from time-series imagery is essential for high-throughput phenotyping, yet it remains computationally challenging due to non-rigid growth dynamics and severe identity fragmentation within entangled canopies. To overcome these stage-dependent ambiguities, we propose ST-DETrack, a spatiotemporal-fusion dual-decoder network designed to preserve branch identity from budding to flowering. Our architecture integrates a spatial decoder, which leverages geometric priors such as position and angle for early-stage tracking, with a temporal decoder that exploits motion consistency to resolve late-stage occlusions. Crucially, an adaptive gating mechanism dynamically shifts reliance between these spatial and temporal cues, while a biological constraint based on negative gravitropism mitigates vertical growth ambiguities. Validated on a Brassica napus dataset, ST-DETrack achieves a Branch Matching Accuracy (BMA) of 93.6%, significantly outperforming spatial and temporal baselines by 28.9 and 3.3 percentage points, respectively. These results demonstrate the method's robustness in maintaining long-term identity consistency amidst complex, dynamic plant architectures.
FloraForge: LLM-Assisted Procedural Generation of Editable and Analysis-Ready 3D Plant Geometric Models For Agricultural Applications
Dec 11, 2025



Accurate 3D plant models are crucial for computational phenotyping and physics-based simulation; however, current approaches face significant limitations. Learning-based reconstruction methods require extensive species-specific training data and lack editability. Procedural modeling offers parametric control but demands specialized expertise in geometric modeling and an in-depth understanding of complex procedural rules, making it inaccessible to domain scientists. We present FloraForge, an LLM-assisted framework that enables domain experts to generate biologically accurate, fully parametric 3D plant models through iterative natural language Plant Refinements (PR), minimizing programming expertise. Our framework leverages LLM-enabled co-design to refine Python scripts that generate parameterized plant geometries as hierarchical B-spline surface representations with botanical constraints with explicit control points and parametric deformation functions. This representation can be easily tessellated into polygonal meshes with arbitrary precision, ensuring compatibility with functional structural plant analysis workflows such as light simulation, computational fluid dynamics, and finite element analysis. We demonstrate the framework on maize, soybean, and mung bean, fitting procedural models to empirical point cloud data through manual refinement of the Plant Descriptor (PD), human-readable files. The pipeline generates dual outputs: triangular meshes for visualization and triangular meshes with additional parametric metadata for quantitative analysis. This approach uniquely combines LLM-assisted template creation, mathematically continuous representations enabling both phenotyping and rendering, and direct parametric control through PD. The framework democratizes sophisticated geometric modeling for plant science while maintaining mathematical rigor.
Data-driven Prediction of Species-Specific Plant Responses to Spectral-Shifting Films from Leaf Phenotypic and Photosynthetic Traits
Nov 19, 2025The application of spectral-shifting films in greenhouses to shift green light to red light has shown variable growth responses across crop species. However, the yield enhancement of crops under altered light quality is related to the collective effects of the specific biophysical characteristics of each species. Considering only one attribute of a crop has limitations in understanding the relationship between sunlight quality adjustments and crop growth performance. Therefore, this study aims to comprehensively link multiple plant phenotypic traits and daily light integral considering the physiological responses of crops to their growth outcomes under SF using artificial intelligence. Between 2021 and 2024, various leafy, fruiting, and root crops were grown in greenhouses covered with either PEF or SF, and leaf reflectance, leaf mass per area, chlorophyll content, daily light integral, and light saturation point were measured from the plants cultivated in each condition. 210 data points were collected, but there was insufficient data to train deep learning models, so a variational autoencoder was used for data augmentation. Most crop yields showed an average increase of 22.5% under SF. These data were used to train several models, including logistic regression, decision tree, random forest, XGBoost, and feedforward neural network (FFNN), aiming to binary classify whether there was a significant effect on yield with SF application. The FFNN achieved a high classification accuracy of 91.4% on a test dataset that was not used for training. This study provide insight into the complex interactions between leaf phenotypic and photosynthetic traits, environmental conditions, and solar spectral components by improving the ability to predict solar spectral shift effects using SF.
ViewSparsifier: Killing Redundancy in Multi-View Plant Phenotyping
Sep 10, 2025Plant phenotyping involves analyzing observable characteristics of plants to better understand their growth, health, and development. In the context of deep learning, this analysis is often approached through single-view classification or regression models. However, these methods often fail to capture all information required for accurate estimation of target phenotypic traits, which can adversely affect plant health assessment and harvest readiness prediction. To address this, the Growth Modelling (GroMo) Grand Challenge at ACM Multimedia 2025 provides a multi-view dataset featuring multiple plants and two tasks: Plant Age Prediction and Leaf Count Estimation. Each plant is photographed from multiple heights and angles, leading to significant overlap and redundancy in the captured information. To learn view-invariant embeddings, we incorporate 24 views, referred to as the selection vector, in a random selection. Our ViewSparsifier approach won both tasks. For further improvement and as a direction for future research, we also experimented with randomized view selection across all five height levels (120 views total), referred to as selection matrices.
3D Plant Root Skeleton Detection and Extraction
Aug 11, 2025Plant roots typically exhibit a highly complex and dense architecture, incorporating numerous slender lateral roots and branches, which significantly hinders the precise capture and modeling of the entire root system. Additionally, roots often lack sufficient texture and color information, making it difficult to identify and track root traits using visual methods. Previous research on roots has been largely confined to 2D studies; however, exploring the 3D architecture of roots is crucial in botany. Since roots grow in real 3D space, 3D phenotypic information is more critical for studying genetic traits and their impact on root development. We have introduced a 3D root skeleton extraction method that efficiently derives the 3D architecture of plant roots from a few images. This method includes the detection and matching of lateral roots, triangulation to extract the skeletal structure of lateral roots, and the integration of lateral and primary roots. We developed a highly complex root dataset and tested our method on it. The extracted 3D root skeletons showed considerable similarity to the ground truth, validating the effectiveness of the model. This method can play a significant role in automated breeding robots. Through precise 3D root structure analysis, breeding robots can better identify plant phenotypic traits, especially root structure and growth patterns, helping practitioners select seeds with superior root systems. This automated approach not only improves breeding efficiency but also reduces manual intervention, making the breeding process more intelligent and efficient, thus advancing modern agriculture.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge