Zixian Su
Decentralizing Test-time Adaptation under Heterogeneous Data Streams
Nov 16, 2024



Abstract:While Test-Time Adaptation (TTA) has shown promise in addressing distribution shifts between training and testing data, its effectiveness diminishes with heterogeneous data streams due to uniform target estimation. As previous attempts merely stabilize model fine-tuning over time to handle continually changing environments, they fundamentally assume a homogeneous target domain at any moment, leaving the intrinsic real-world data heterogeneity unresolved. This paper delves into TTA under heterogeneous data streams, moving beyond current model-centric limitations. By revisiting TTA from a data-centric perspective, we discover that decomposing samples into Fourier space facilitates an accurate data separation across different frequency levels. Drawing from this insight, we propose a novel Frequency-based Decentralized Adaptation (FreDA) framework, which transitions data from globally heterogeneous to locally homogeneous in Fourier space and employs decentralized adaptation to manage diverse distribution shifts.Interestingly, we devise a novel Fourier-based augmentation strategy to assist in decentralizing adaptation, which individually enhances sample quality for capturing each type of distribution shifts. Extensive experiments across various settings (corrupted, natural, and medical environments) demonstrate the superiority of our proposed framework over the state-of-the-arts.
SCMix: Stochastic Compound Mixing for Open Compound Domain Adaptation in Semantic Segmentation
May 23, 2024Abstract:Open compound domain adaptation (OCDA) aims to transfer knowledge from a labeled source domain to a mix of unlabeled homogeneous compound target domains while generalizing to open unseen domains. Existing OCDA methods solve the intra-domain gaps by a divide-and-conquer strategy, which divides the problem into several individual and parallel domain adaptation (DA) tasks. Such approaches often contain multiple sub-networks or stages, which may constrain the model's performance. In this work, starting from the general DA theory, we establish the generalization bound for the setting of OCDA. Built upon this, we argue that conventional OCDA approaches may substantially underestimate the inherent variance inside the compound target domains for model generalization. We subsequently present Stochastic Compound Mixing (SCMix), an augmentation strategy with the primary objective of mitigating the divergence between source and mixed target distributions. We provide theoretical analysis to substantiate the superiority of SCMix and prove that the previous methods are sub-groups of our methods. Extensive experiments show that our method attains a lower empirical risk on OCDA semantic segmentation tasks, thus supporting our theories. Combining the transformer architecture, SCMix achieves a notable performance boost compared to the SoTA results.
Rethinking Spectral Graph Neural Networks with Spatially Adaptive Filtering
Jan 30, 2024Abstract:Whilst spectral Graph Neural Networks (GNNs) are theoretically well-founded in the spectral domain, their practical reliance on polynomial approximation implies a profound linkage to the spatial domain. As previous studies rarely examine spectral GNNs from the spatial perspective, their spatial-domain interpretability remains elusive, e.g., what information is essentially encoded by spectral GNNs in the spatial domain? In this paper, to answer this question, we establish a theoretical connection between spectral filtering and spatial aggregation, unveiling an intrinsic interaction that spectral filtering implicitly leads the original graph to an adapted new graph, explicitly computed for spatial aggregation. Both theoretical and empirical investigations reveal that the adapted new graph not only exhibits non-locality but also accommodates signed edge weights to reflect label consistency among nodes. These findings thus highlight the interpretable role of spectral GNNs in the spatial domain and inspire us to rethink graph spectral filters beyond the fixed-order polynomials, which neglect global information. Built upon the theoretical findings, we revisit the state-of-the-art spectral GNNs and propose a novel Spatially Adaptive Filtering (SAF) framework, which leverages the adapted new graph by spectral filtering for an auxiliary non-local aggregation. Notably, our proposed SAF comprehensively models both node similarity and dissimilarity from a global perspective, therefore alleviating persistent deficiencies of GNNs related to long-range dependencies and graph heterophily. Extensive experiments over 13 node classification benchmarks demonstrate the superiority of our proposed framework to the state-of-the-art models.
Unraveling Batch Normalization for Realistic Test-Time Adaptation
Dec 15, 2023



Abstract:While recent test-time adaptations exhibit efficacy by adjusting batch normalization to narrow domain disparities, their effectiveness diminishes with realistic mini-batches due to inaccurate target estimation. As previous attempts merely introduce source statistics to mitigate this issue, the fundamental problem of inaccurate target estimation still persists, leaving the intrinsic test-time domain shifts unresolved. This paper delves into the problem of mini-batch degradation. By unraveling batch normalization, we discover that the inexact target statistics largely stem from the substantially reduced class diversity in batch. Drawing upon this insight, we introduce a straightforward tool, Test-time Exponential Moving Average (TEMA), to bridge the class diversity gap between training and testing batches. Importantly, our TEMA adaptively extends the scope of typical methods beyond the current batch to incorporate a diverse set of class information, which in turn boosts an accurate target estimation. Built upon this foundation, we further design a novel layer-wise rectification strategy to consistently promote test-time performance. Our proposed method enjoys a unique advantage as it requires neither training nor tuning parameters, offering a truly hassle-free solution. It significantly enhances model robustness against shifted domains and maintains resilience in diverse real-world scenarios with various batch sizes, achieving state-of-the-art performance on several major benchmarks. Code is available at \url{https://github.com/kiwi12138/RealisticTTA}.
Rethinking Data Augmentation for Single-source Domain Generalization in Medical Image Segmentation
Nov 27, 2022Abstract:Single-source domain generalization (SDG) in medical image segmentation is a challenging yet essential task as domain shifts are quite common among clinical image datasets. Previous attempts most conduct global-only/random augmentation. Their augmented samples are usually insufficient in diversity and informativeness, thus failing to cover the possible target domain distribution. In this paper, we rethink the data augmentation strategy for SDG in medical image segmentation. Motivated by the class-level representation invariance and style mutability of medical images, we hypothesize that unseen target data can be sampled from a linear combination of $C$ (the class number) random variables, where each variable follows a location-scale distribution at the class level. Accordingly, data augmented can be readily made by sampling the random variables through a general form. On the empirical front, we implement such strategy with constrained B$\acute{\rm e}$zier transformation on both global and local (i.e. class-level) regions, which can largely increase the augmentation diversity. A Saliency-balancing Fusion mechanism is further proposed to enrich the informativeness by engaging the gradient information, guiding augmentation with proper orientation and magnitude. As an important contribution, we prove theoretically that our proposed augmentation can lead to an upper bound of the generalization risk on the unseen target domain, thus confirming our hypothesis. Combining the two strategies, our Saliency-balancing Location-scale Augmentation (SLAug) exceeds the state-of-the-art works by a large margin in two challenging SDG tasks. Code is available at https://github.com/Kaiseem/SLAug .
Mind The Gap: Alleviating Local Imbalance for Unsupervised Cross-Modality Medical Image Segmentation
May 24, 2022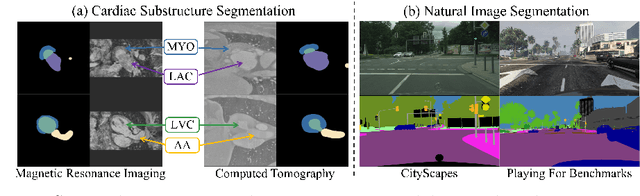
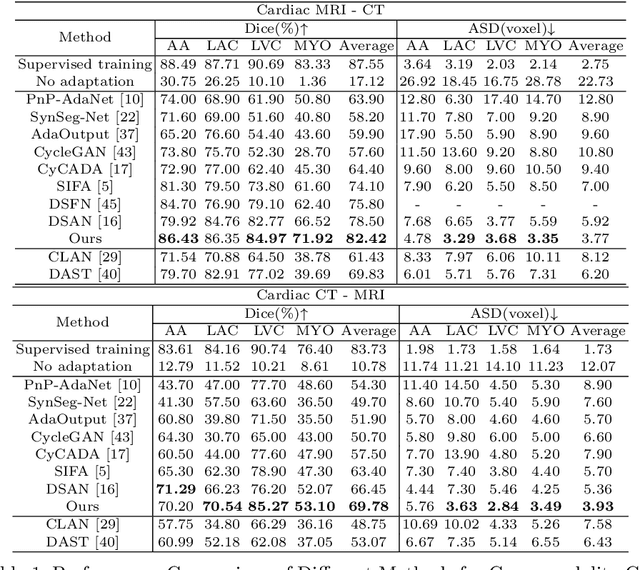
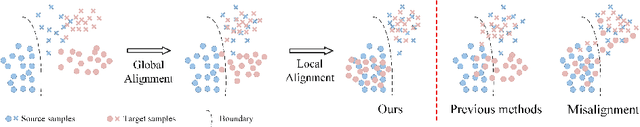
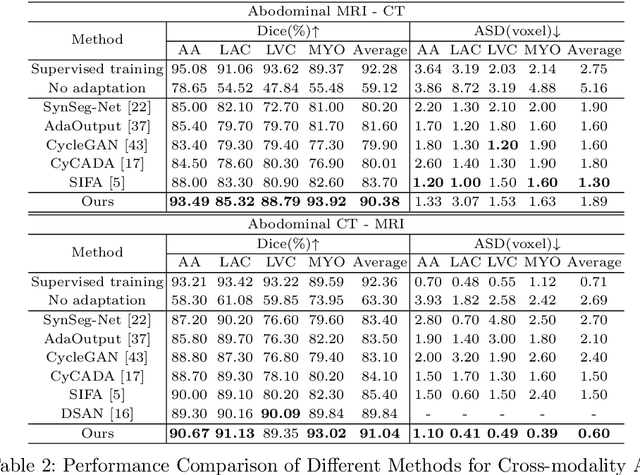
Abstract:Unsupervised cross-modality medical image adaptation aims to alleviate the severe domain gap between different imaging modalities without using the target domain label. A key in this campaign relies upon aligning the distributions of source and target domain. One common attempt is to enforce the global alignment between two domains, which, however, ignores the fatal local-imbalance domain gap problem, i.e., some local features with larger domain gap are harder to transfer. Recently, some methods conduct alignment focusing on local regions to improve the efficiency of model learning. While this operation may cause a deficiency of critical information from contexts. To tackle this limitation, we propose a novel strategy to alleviate the domain gap imbalance considering the characteristics of medical images, namely Global-Local Union Alignment. Specifically, a feature-disentanglement style-transfer module first synthesizes the target-like source-content images to reduce the global domain gap. Then, a local feature mask is integrated to reduce the 'inter-gap' for local features by prioritizing those discriminative features with larger domain gap. This combination of global and local alignment can precisely localize the crucial regions in segmentation target while preserving the overall semantic consistency. We conduct a series of experiments with two cross-modality adaptation tasks, i,e. cardiac substructure and abdominal multi-organ segmentation. Experimental results indicate that our method exceeds the SOTA methods by 3.92% Dice score in MRI-CT cardiac segmentation and 3.33% in the reverse direction.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022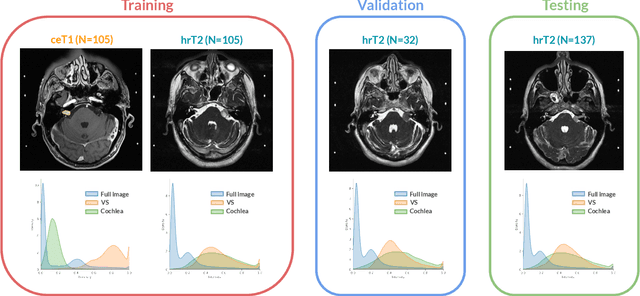
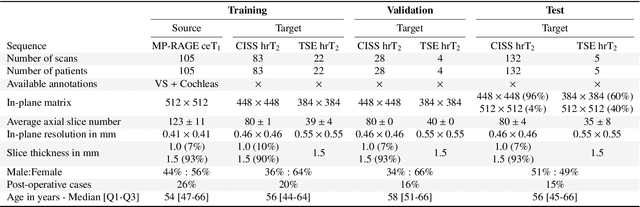
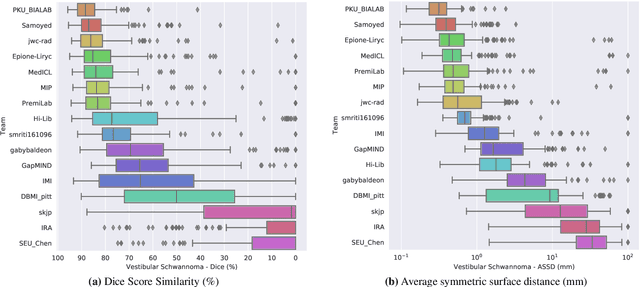
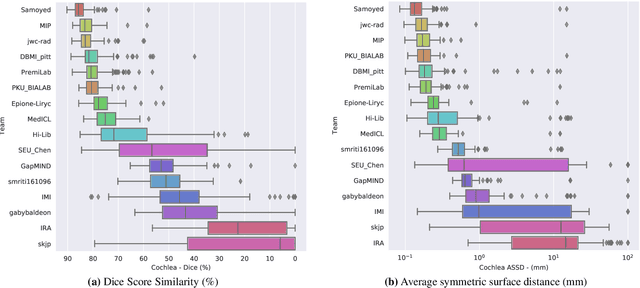
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge