Ziliang Li
Department of Electrical and Computer Engineering, University of Toronto, Toronto, ON, Canada
IRS: Instance-Level 3D Scene Graphs via Room Prior Guided LiDAR-Camera Fusion
Jun 07, 2025Abstract:Indoor scene understanding remains a fundamental challenge in robotics, with direct implications for downstream tasks such as navigation and manipulation. Traditional approaches often rely on closed-set recognition or loop closure, limiting their adaptability in open-world environments. With the advent of visual foundation models (VFMs), open-vocabulary recognition and natural language querying have become feasible, unlocking new possibilities for 3D scene graph construction. In this paper, we propose a robust and efficient framework for instance-level 3D scene graph construction via LiDAR-camera fusion. Leveraging LiDAR's wide field of view (FOV) and long-range sensing capabilities, we rapidly acquire room-level geometric priors. Multi-level VFMs are employed to improve the accuracy and consistency of semantic extraction. During instance fusion, room-based segmentation enables parallel processing, while the integration of geometric and semantic cues significantly enhances fusion accuracy and robustness. Compared to state-of-the-art methods, our approach achieves up to an order-of-magnitude improvement in construction speed while maintaining high semantic precision. Extensive experiments in both simulated and real-world environments validate the effectiveness of our approach. We further demonstrate its practical value through a language-guided semantic navigation task, highlighting its potential for real-world robotic applications.
CandidateDrug4Cancer: An Open Molecular Graph Learning Benchmark on Drug Discovery for Cancer
Mar 02, 2022
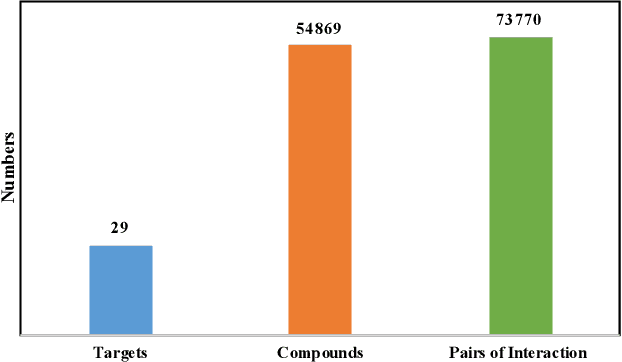
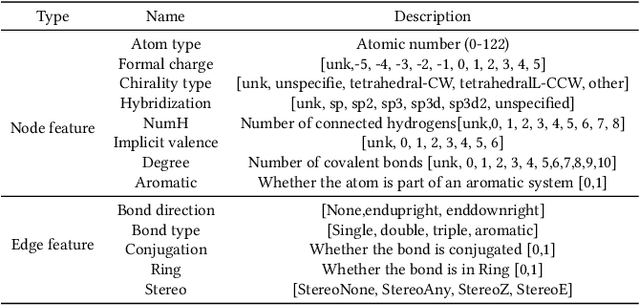
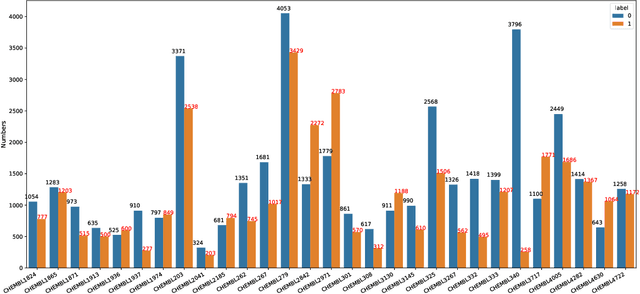
Abstract:Anti-cancer drug discoveries have been serendipitous, we sought to present the Open Molecular Graph Learning Benchmark, named CandidateDrug4Cancer, a challenging and realistic benchmark dataset to facilitate scalable, robust, and reproducible graph machine learning research for anti-cancer drug discovery. CandidateDrug4Cancer dataset encompasses multiple most-mentioned 29 targets for cancer, covering 54869 cancer-related drug molecules which are ranged from pre-clinical, clinical and FDA-approved. Besides building the datasets, we also perform benchmark experiments with effective Drug Target Interaction (DTI) prediction baselines using descriptors and expressive graph neural networks. Experimental results suggest that CandidateDrug4Cancer presents significant challenges for learning molecular graphs and targets in practical application, indicating opportunities for future researches on developing candidate drugs for treating cancers.
Pairwise Half-graph Discrimination: A Simple Graph-level Self-supervised Strategy for Pre-training Graph Neural Networks
Oct 26, 2021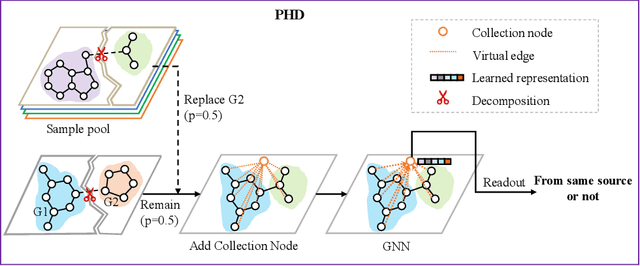
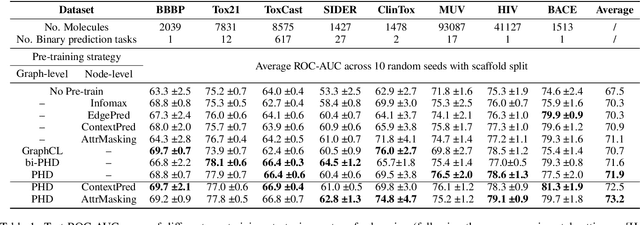
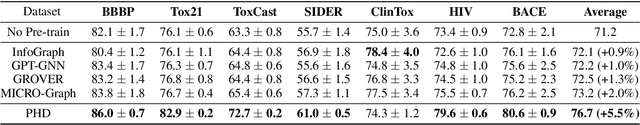
Abstract:Self-supervised learning has gradually emerged as a powerful technique for graph representation learning. However, transferable, generalizable, and robust representation learning on graph data still remains a challenge for pre-training graph neural networks. In this paper, we propose a simple and effective self-supervised pre-training strategy, named Pairwise Half-graph Discrimination (PHD), that explicitly pre-trains a graph neural network at graph-level. PHD is designed as a simple binary classification task to discriminate whether two half-graphs come from the same source. Experiments demonstrate that the PHD is an effective pre-training strategy that offers comparable or superior performance on 13 graph classification tasks compared with state-of-the-art strategies, and achieves notable improvements when combined with node-level strategies. Moreover, the visualization of learned representation revealed that PHD strategy indeed empowers the model to learn graph-level knowledge like the molecular scaffold. These results have established PHD as a powerful and effective self-supervised learning strategy in graph-level representation learning.
Winner Team Mia at TextVQA Challenge 2021: Vision-and-Language Representation Learning with Pre-trained Sequence-to-Sequence Model
Jun 24, 2021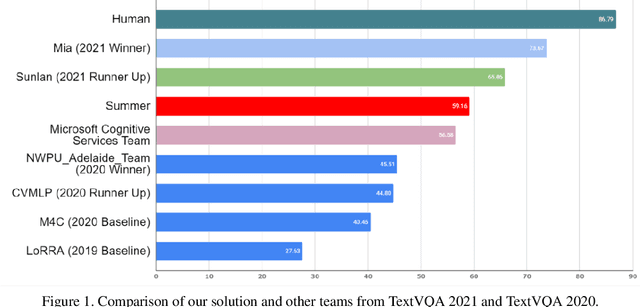
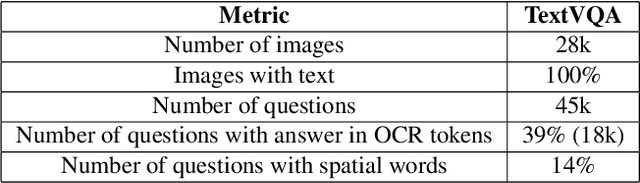
Abstract:TextVQA requires models to read and reason about text in images to answer questions about them. Specifically, models need to incorporate a new modality of text present in the images and reason over it to answer TextVQA questions. In this challenge, we use generative model T5 for TextVQA task. Based on pre-trained checkpoint T5-3B from HuggingFace repository, two other pre-training tasks including masked language modeling(MLM) and relative position prediction(RPP) are designed to better align object feature and scene text. In the stage of pre-training, encoder is dedicate to handle the fusion among multiple modalities: question text, object text labels, scene text labels, object visual features, scene visual features. After that decoder generates the text sequence step-by-step, cross entropy loss is required by default. We use a large-scale scene text dataset in pre-training and then fine-tune the T5-3B with the TextVQA dataset only.
Interpretable discovery of new semiconductors with machine learning
Jan 12, 2021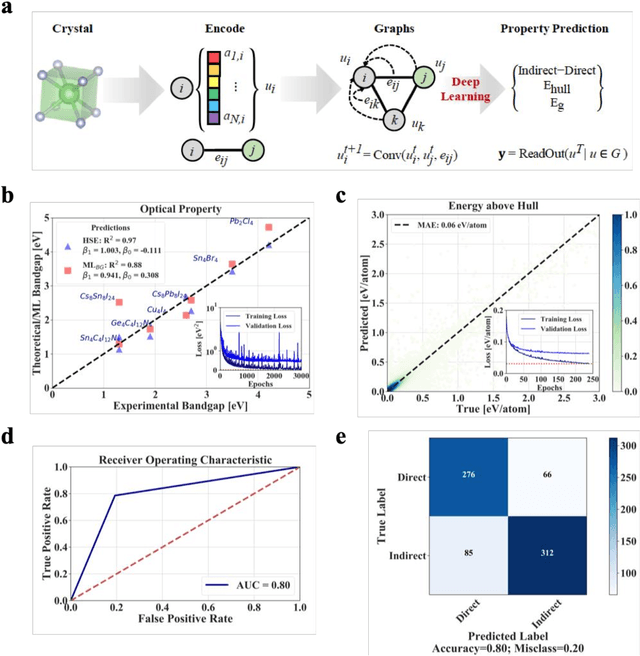
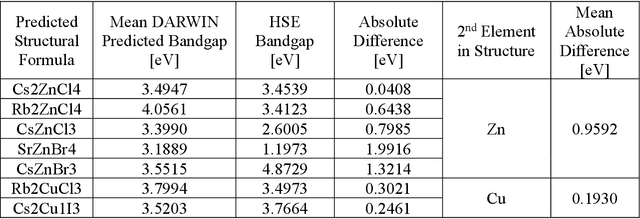
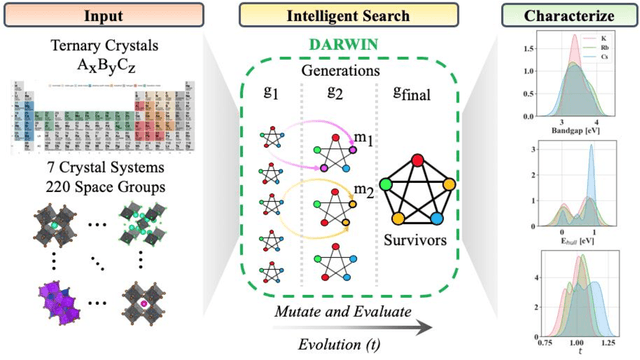
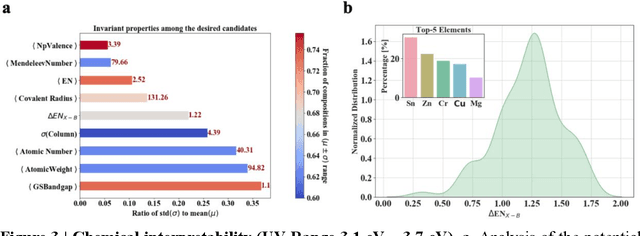
Abstract:Machine learning models of materials$^{1-5}$ accelerate discovery compared to ab initio methods: deep learning models now reproduce density functional theory (DFT)-calculated results at one hundred thousandths of the cost of DFT$^{6}$. To provide guidance in experimental materials synthesis, these need to be coupled with an accurate yet effective search algorithm and training data consistent with experimental observations. Here we report an evolutionary algorithm powered search which uses machine-learned surrogate models trained on high-throughput hybrid functional DFT data benchmarked against experimental bandgaps: Deep Adaptive Regressive Weighted Intelligent Network (DARWIN). The strategy enables efficient search over the materials space of ~10$^8$ ternaries and 10$^{11}$ quaternaries$^{7}$ for candidates with target properties. It provides interpretable design rules, such as our finding that the difference in the electronegativity between the halide and B-site cation being a strong predictor of ternary structural stability. As an example, when we seek UV emission, DARWIN predicts K$_2$CuX$_3$ (X = Cl, Br) as a promising materials family, based on its electronegativity difference. We synthesized and found these materials to be stable, direct bandgap UV emitters. The approach also allows knowledge distillation for use by humans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge