Yawen Wu
FedCoCo: A Memory Efficient Federated Self-supervised Framework for On-Device Visual Representation Learning
Dec 02, 2022



Abstract:The ubiquity of edge devices has led to a growing amount of unlabeled data produced at the edge. Deep learning models deployed on edge devices are required to learn from these unlabeled data to continuously improve accuracy. Self-supervised representation learning has achieved promising performances using centralized unlabeled data. However, the increasing awareness of privacy protection limits centralizing the distributed unlabeled image data on edge devices. While federated learning has been widely adopted to enable distributed machine learning with privacy preservation, without a data selection method to efficiently select streaming data, the traditional federated learning framework fails to handle these huge amounts of decentralized unlabeled data with limited storage resources on edge. To address these challenges, we propose a Federated on-device Contrastive learning framework with Coreset selection, which we call FedCoCo, to automatically select a coreset that consists of the most representative samples into the replay buffer on each device. It preserves data privacy as each client does not share raw data while learning good visual representations. Experiments demonstrate the effectiveness and significance of the proposed method in visual representation learning.
Enabling Weakly-Supervised Temporal Action Localization from On-Device Learning of the Video Stream
Aug 25, 2022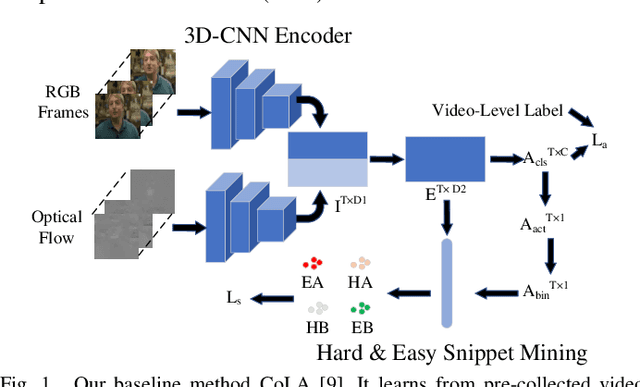
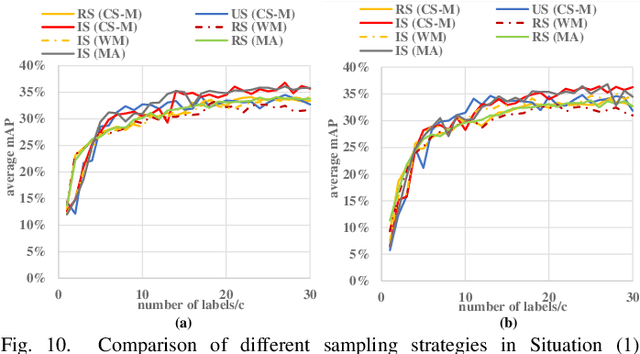
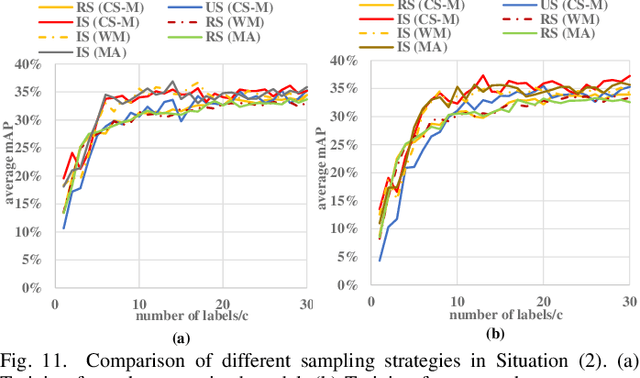
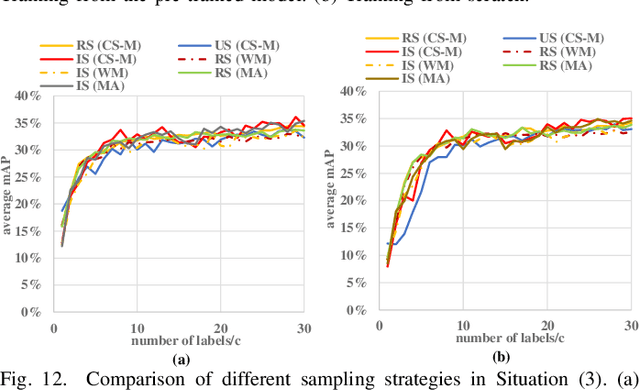
Abstract:Detecting actions in videos have been widely applied in on-device applications. Practical on-device videos are always untrimmed with both action and background. It is desirable for a model to both recognize the class of action and localize the temporal position where the action happens. Such a task is called temporal action location (TAL), which is always trained on the cloud where multiple untrimmed videos are collected and labeled. It is desirable for a TAL model to continuously and locally learn from new data, which can directly improve the action detection precision while protecting customers' privacy. However, it is non-trivial to train a TAL model, since tremendous video samples with temporal annotations are required. However, annotating videos frame by frame is exorbitantly time-consuming and expensive. Although weakly-supervised TAL (W-TAL) has been proposed to learn from untrimmed videos with only video-level labels, such an approach is also not suitable for on-device learning scenarios. In practical on-device learning applications, data are collected in streaming. Dividing such a long video stream into multiple video segments requires lots of human effort, which hinders the exploration of applying the TAL tasks to realistic on-device learning applications. To enable W-TAL models to learn from a long, untrimmed streaming video, we propose an efficient video learning approach that can directly adapt to new environments. We first propose a self-adaptive video dividing approach with a contrast score-based segment merging approach to convert the video stream into multiple segments. Then, we explore different sampling strategies on the TAL tasks to request as few labels as possible. To the best of our knowledge, we are the first attempt to directly learn from the on-device, long video stream.
Federated Self-Supervised Contrastive Learning and Masked Autoencoder for Dermatological Disease Diagnosis
Aug 24, 2022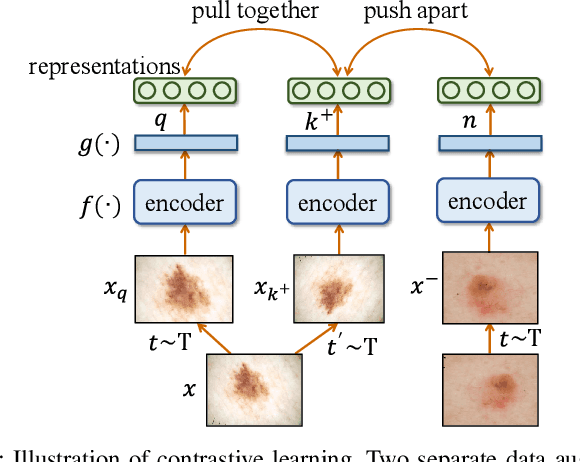
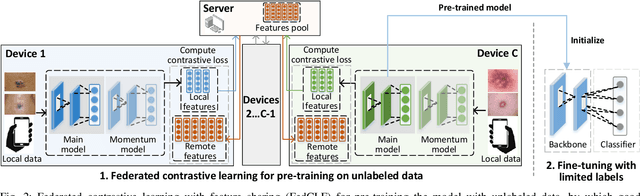
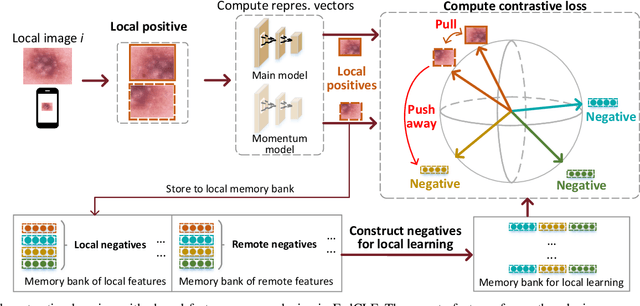
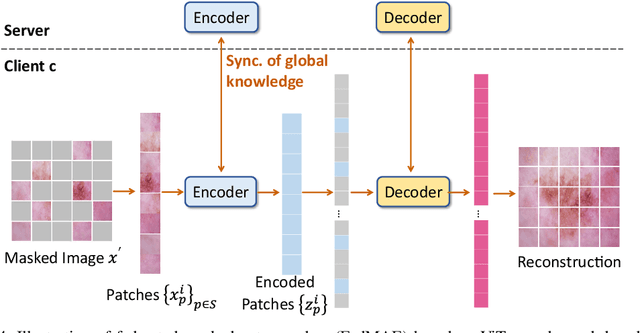
Abstract:In dermatological disease diagnosis, the private data collected by mobile dermatology assistants exist on distributed mobile devices of patients. Federated learning (FL) can use decentralized data to train models while keeping data local. Existing FL methods assume all the data have labels. However, medical data often comes without full labels due to high labeling costs. Self-supervised learning (SSL) methods, contrastive learning (CL) and masked autoencoders (MAE), can leverage the unlabeled data to pre-train models, followed by fine-tuning with limited labels. However, combining SSL and FL has unique challenges. For example, CL requires diverse data but each device only has limited data. For MAE, while Vision Transformer (ViT) based MAE has higher accuracy over CNNs in centralized learning, MAE's performance in FL with unlabeled data has not been investigated. Besides, the ViT synchronization between the server and clients is different from traditional CNNs. Therefore, special synchronization methods need to be designed. In this work, we propose two federated self-supervised learning frameworks for dermatological disease diagnosis with limited labels. The first one features lower computation costs, suitable for mobile devices. The second one features high accuracy and fits high-performance servers. Based on CL, we proposed federated contrastive learning with feature sharing (FedCLF). Features are shared for diverse contrastive information without sharing raw data for privacy. Based on MAE, we proposed FedMAE. Knowledge split separates the global and local knowledge learned from each client. Only global knowledge is aggregated for higher generalization performance. Experiments on dermatological disease datasets show superior accuracy of the proposed frameworks over state-of-the-arts.
Achieving Fairness in Dermatological Disease Diagnosis through Automatic Weight Adjusting Federated Learning and Personalization
Aug 23, 2022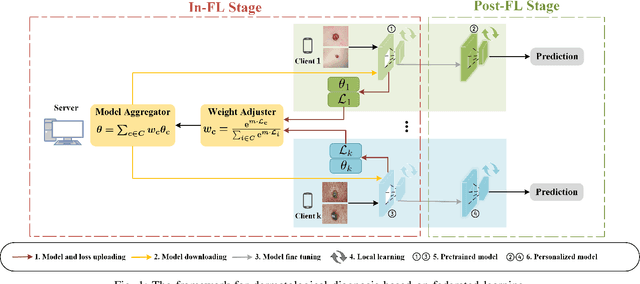
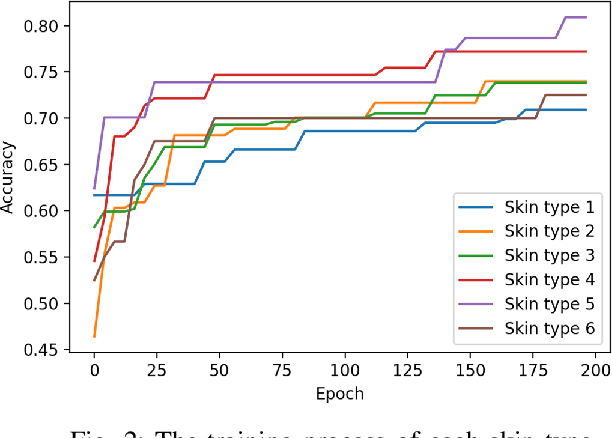
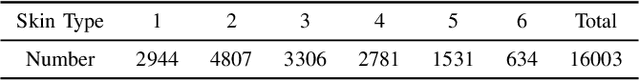
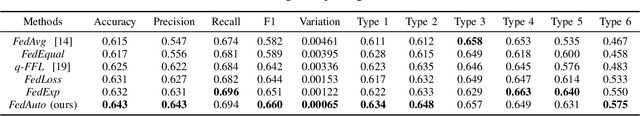
Abstract:Dermatological diseases pose a major threat to the global health, affecting almost one-third of the world's population. Various studies have demonstrated that early diagnosis and intervention are often critical to prognosis and outcome. To this end, the past decade has witnessed the rapid evolvement of deep learning based smartphone apps, which allow users to conveniently and timely identify issues that have emerged around their skins. In order to collect sufficient data needed by deep learning and at the same time protect patient privacy, federated learning is often used, where individual clients aggregate a global model while keeping datasets local. However, existing federated learning frameworks are mostly designed to optimize the overall performance, while common dermatological datasets are heavily imbalanced. When applying federated learning to such datasets, significant disparities in diagnosis accuracy may occur. To address such a fairness issue, this paper proposes a fairness-aware federated learning framework for dermatological disease diagnosis. The framework is divided into two stages: In the first in-FL stage, clients with different skin types are trained in a federated learning process to construct a global model for all skin types. An automatic weight aggregator is used in this process to assign higher weights to the client with higher loss, and the intensity of the aggregator is determined by the level of difference between losses. In the latter post-FL stage, each client fine-tune its personalized model based on the global model in the in-FL stage. To achieve better fairness, models from different epochs are selected for each client to keep the accuracy difference of different skin types within 0.05. Experiments indicate that our proposed framework effectively improves both fairness and accuracy compared with the state-of-the-art.
Distributed Contrastive Learning for Medical Image Segmentation
Aug 07, 2022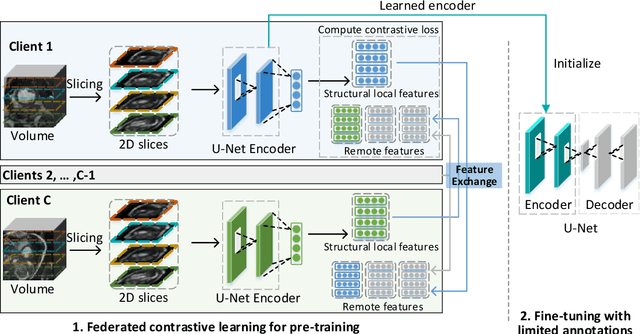
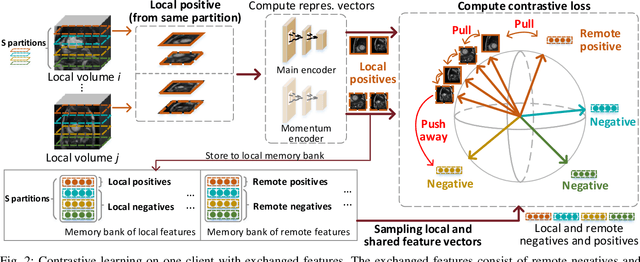
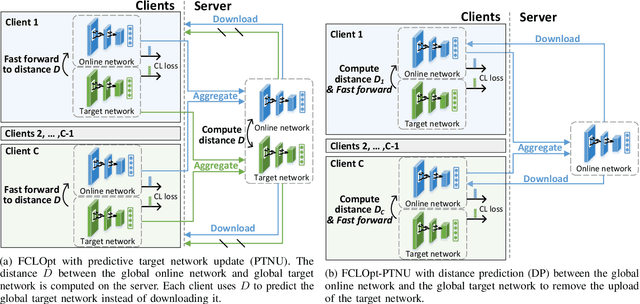
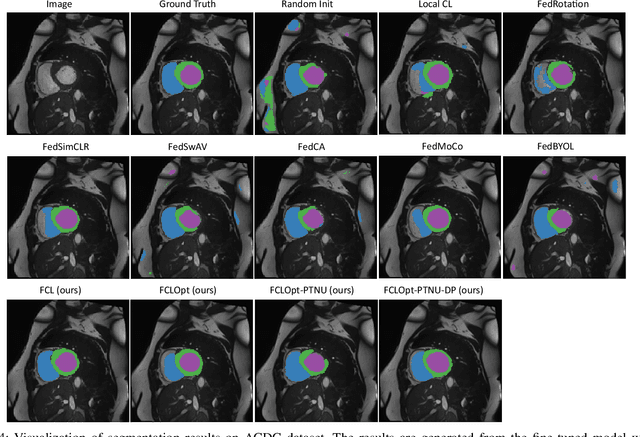
Abstract:Supervised deep learning needs a large amount of labeled data to achieve high performance. However, in medical imaging analysis, each site may only have a limited amount of data and labels, which makes learning ineffective. Federated learning (FL) can learn a shared model from decentralized data. But traditional FL requires fully-labeled data for training, which is very expensive to obtain. Self-supervised contrastive learning (CL) can learn from unlabeled data for pre-training, followed by fine-tuning with limited annotations. However, when adopting CL in FL, the limited data diversity on each site makes federated contrastive learning (FCL) ineffective. In this work, we propose two federated self-supervised learning frameworks for volumetric medical image segmentation with limited annotations. The first one features high accuracy and fits high-performance servers with high-speed connections. The second one features lower communication costs, suitable for mobile devices. In the first framework, features are exchanged during FCL to provide diverse contrastive data to each site for effective local CL while keeping raw data private. Global structural matching aligns local and remote features for a unified feature space among different sites. In the second framework, to reduce the communication cost for feature exchanging, we propose an optimized method FCLOpt that does not rely on negative samples. To reduce the communications of model download, we propose the predictive target network update (PTNU) that predicts the parameters of the target network. Based on PTNU, we propose the distance prediction (DP) to remove most of the uploads of the target network. Experiments on a cardiac MRI dataset show the proposed two frameworks substantially improve the segmentation and generalization performance compared with state-of-the-art techniques.
Federated Contrastive Learning for Volumetric Medical Image Segmentation
Apr 23, 2022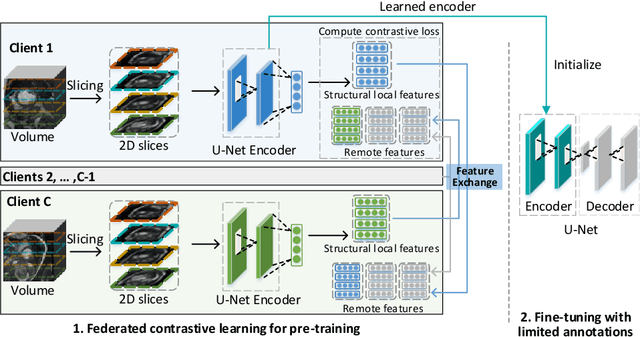
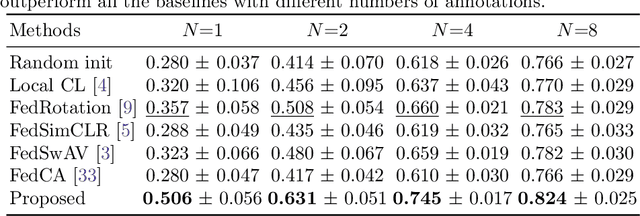
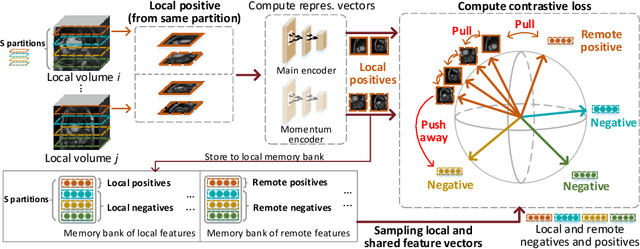
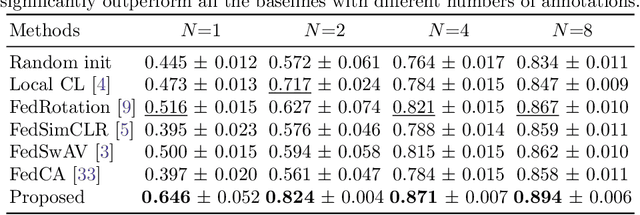
Abstract:Supervised deep learning needs a large amount of labeled data to achieve high performance. However, in medical imaging analysis, each site may only have a limited amount of data and labels, which makes learning ineffective. Federated learning (FL) can help in this regard by learning a shared model while keeping training data local for privacy. Traditional FL requires fully-labeled data for training, which is inconvenient or sometimes infeasible to obtain due to high labeling cost and the requirement of expertise. Contrastive learning (CL), as a self-supervised learning approach, can effectively learn from unlabeled data to pre-train a neural network encoder, followed by fine-tuning for downstream tasks with limited annotations. However, when adopting CL in FL, the limited data diversity on each client makes federated contrastive learning (FCL) ineffective. In this work, we propose an FCL framework for volumetric medical image segmentation with limited annotations. More specifically, we exchange the features in the FCL pre-training process such that diverse contrastive data are provided to each site for effective local CL while keeping raw data private. Based on the exchanged features, global structural matching further leverages the structural similarity to align local features to the remote ones such that a unified feature space can be learned among different sites. Experiments on a cardiac MRI dataset show the proposed framework substantially improves the segmentation performance compared with state-of-the-art techniques.
FairPrune: Achieving Fairness Through Pruning for Dermatological Disease Diagnosis
Mar 04, 2022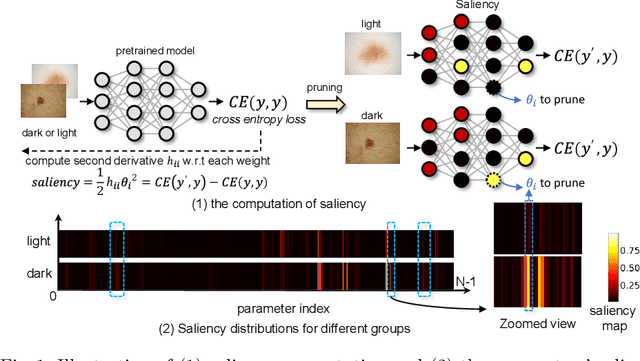
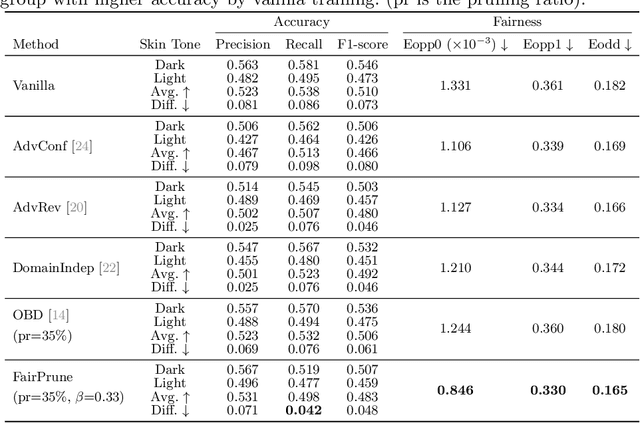
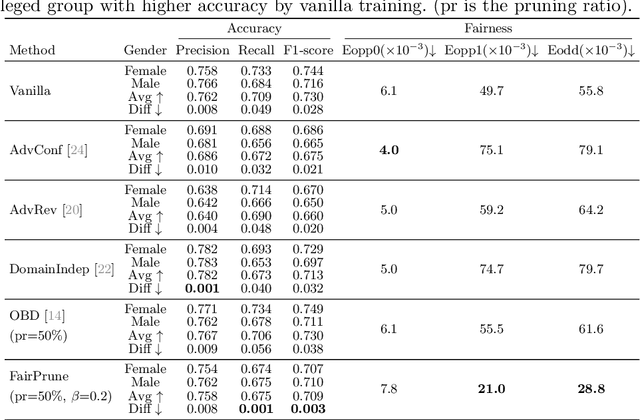
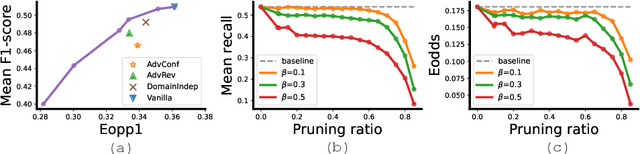
Abstract:Many works have shown that deep learning-based medical image classification models can exhibit bias toward certain demographic attributes like race, gender, and age. Existing bias mitigation methods primarily focus on learning debiased models, which may not necessarily guarantee all sensitive information can be removed and usually comes with considerable accuracy degradation on both privileged and unprivileged groups. To tackle this issue, we propose a method, FairPrune, that achieves fairness by pruning. Conventionally, pruning is used to reduce the model size for efficient inference. However, we show that pruning can also be a powerful tool to achieve fairness. Our observation is that during pruning, each parameter in the model has different importance for different groups' accuracy. By pruning the parameters based on this importance difference, we can reduce the accuracy difference between the privileged group and the unprivileged group to improve fairness without a large accuracy drop. To this end, we use the second derivative of the parameters of a pre-trained model to quantify the importance of each parameter with respect to the model accuracy for each group. Experiments on two skin lesion diagnosis datasets over multiple sensitive attributes demonstrate that our method can greatly improve fairness while keeping the average accuracy of both groups as high as possible.
The Larger The Fairer? Small Neural Networks Can Achieve Fairness for Edge Devices
Feb 23, 2022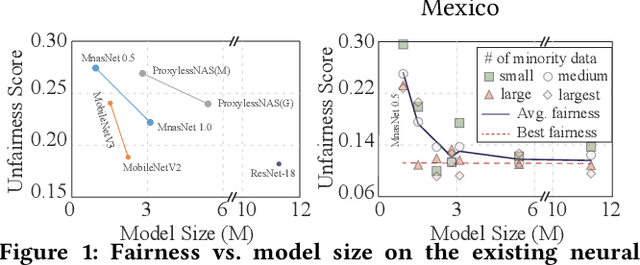
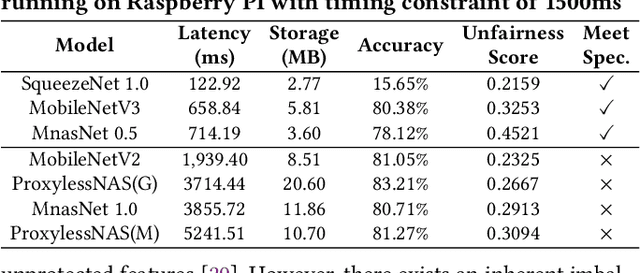
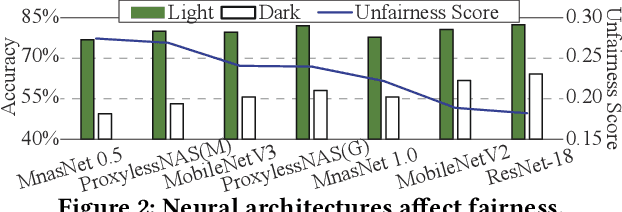
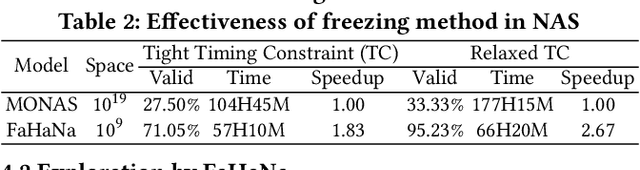
Abstract:Along with the progress of AI democratization, neural networks are being deployed more frequently in edge devices for a wide range of applications. Fairness concerns gradually emerge in many applications, such as face recognition and mobile medical. One fundamental question arises: what will be the fairest neural architecture for edge devices? By examining the existing neural networks, we observe that larger networks typically are fairer. But, edge devices call for smaller neural architectures to meet hardware specifications. To address this challenge, this work proposes a novel Fairness- and Hardware-aware Neural architecture search framework, namely FaHaNa. Coupled with a model freezing approach, FaHaNa can efficiently search for neural networks with balanced fairness and accuracy, while guaranteed to meet hardware specifications. Results show that FaHaNa can identify a series of neural networks with higher fairness and accuracy on a dermatology dataset. Target edge devices, FaHaNa finds a neural architecture with slightly higher accuracy, 5.28x smaller size, 15.14% higher fairness score, compared with MobileNetV2; meanwhile, on Raspberry PI and Odroid XU-4, it achieves 5.75x and 5.79x speedup.
Learn by Challenging Yourself: Contrastive Visual Representation Learning with Hard Sample Generation
Feb 14, 2022Abstract:Contrastive learning (CL), a self-supervised learning approach, can effectively learn visual representations from unlabeled data. However, CL requires learning on vast quantities of diverse data to achieve good performance, without which the performance of CL will greatly degrade. To tackle this problem, we propose a framework with two approaches to improve the data efficiency of CL training by generating beneficial samples and joint learning. The first approach generates hard samples for the main model. The generator is jointly learned with the main model to dynamically customize hard samples based on the training state of the main model. With the progressively growing knowledge of the main model, the generated samples also become harder to constantly encourage the main model to learn better representations. Besides, a pair of data generators are proposed to generate similar but distinct samples as positive pairs. In joint learning, the hardness of a positive pair is progressively increased by decreasing their similarity. In this way, the main model learns to cluster hard positives by pulling the representations of similar yet distinct samples together, by which the representations of similar samples are well-clustered and better representations can be learned. Comprehensive experiments show superior accuracy and data efficiency of the proposed methods over the state-of-the-art on multiple datasets. For example, about 5% accuracy improvement on ImageNet-100 and CIFAR-10, and more than 6% accuracy improvement on CIFAR-100 are achieved for linear classification. Besides, up to 2x data efficiency for linear classification and up to 5x data efficiency for transfer learning are achieved.
Federated Contrastive Learning for Dermatological Disease Diagnosis via On-device Learning
Feb 14, 2022



Abstract:Deep learning models have been deployed in an increasing number of edge and mobile devices to provide healthcare. These models rely on training with a tremendous amount of labeled data to achieve high accuracy. However, for medical applications such as dermatological disease diagnosis, the private data collected by mobile dermatology assistants exist on distributed mobile devices of patients, and each device only has a limited amount of data. Directly learning from limited data greatly deteriorates the performance of learned models. Federated learning (FL) can train models by using data distributed on devices while keeping the data local for privacy. Existing works on FL assume all the data have ground-truth labels. However, medical data often comes without any accompanying labels since labeling requires expertise and results in prohibitively high labor costs. The recently developed self-supervised learning approach, contrastive learning (CL), can leverage the unlabeled data to pre-train a model, after which the model is fine-tuned on limited labeled data for dermatological disease diagnosis. However, simply combining CL with FL as federated contrastive learning (FCL) will result in ineffective learning since CL requires diverse data for learning but each device only has limited data. In this work, we propose an on-device FCL framework for dermatological disease diagnosis with limited labels. Features are shared in the FCL pre-training process to provide diverse and accurate contrastive information. After that, the pre-trained model is fine-tuned with local labeled data independently on each device or collaboratively with supervised federated learning on all devices. Experiments on dermatological disease datasets show that the proposed framework effectively improves the recall and precision of dermatological disease diagnosis compared with state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge