Xiaomei Huang
HoVer-Trans: Anatomy-aware HoVer-Transformer for ROI-free Breast Cancer Diagnosis in Ultrasound Images
May 17, 2022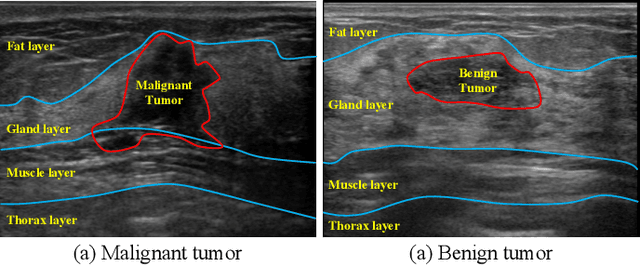
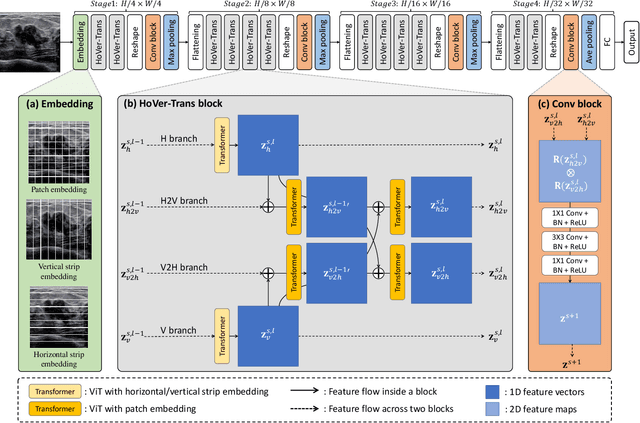
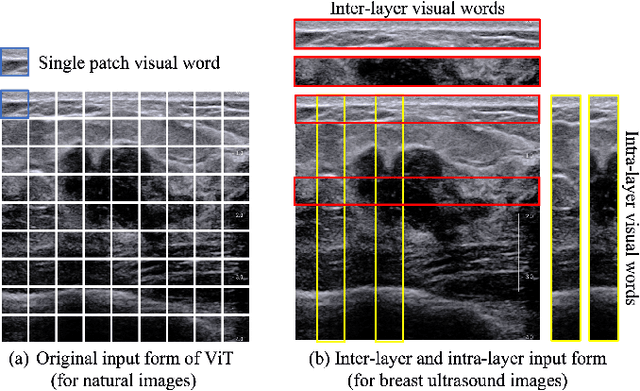
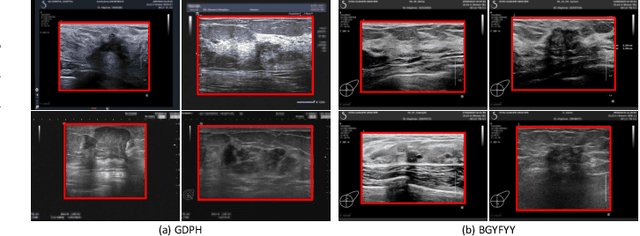
Abstract:Ultrasonography is an important routine examination for breast cancer diagnosis, due to its non-invasive, radiation-free and low-cost properties. However, it is still not the first-line screening test for breast cancer due to its inherent limitations. It would be a tremendous success if we can precisely diagnose breast cancer by breast ultrasound images (BUS). Many learning-based computer-aided diagnostic methods have been proposed to achieve breast cancer diagnosis/lesion classification. However, most of them require a pre-define ROI and then classify the lesion inside the ROI. Conventional classification backbones, such as VGG16 and ResNet50, can achieve promising classification results with no ROI requirement. But these models lack interpretability, thus restricting their use in clinical practice. In this study, we propose a novel ROI-free model for breast cancer diagnosis in ultrasound images with interpretable feature representations. We leverage the anatomical prior knowledge that malignant and benign tumors have different spatial relationships between different tissue layers, and propose a HoVer-Transformer to formulate this prior knowledge. The proposed HoVer-Trans block extracts the inter- and intra-layer spatial information horizontally and vertically. We conduct and release an open dataset GDPH&GYFYY for breast cancer diagnosis in BUS. The proposed model is evaluated in three datasets by comparing with four CNN-based models and two vision transformer models via a five-fold cross validation. It achieves state-of-the-art classification performance with the best model interpretability.
Multi-Layer Pseudo-Supervision for Histopathology Tissue Semantic Segmentation using Patch-level Classification Labels
Oct 14, 2021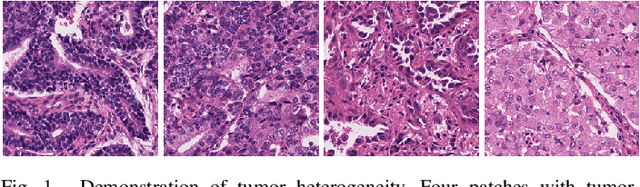

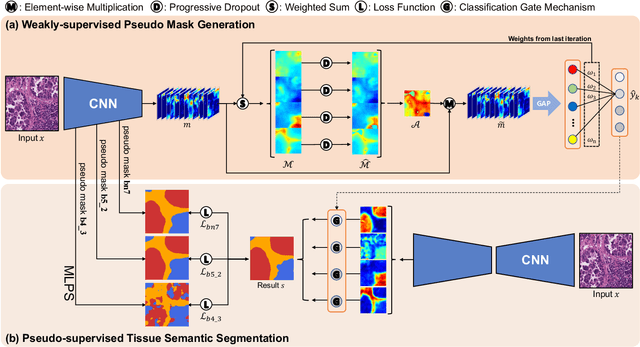
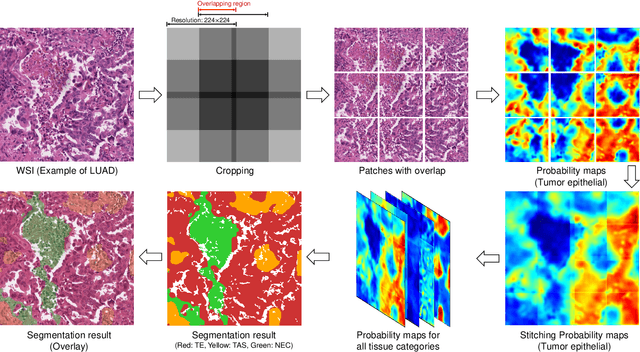
Abstract:Tissue-level semantic segmentation is a vital step in computational pathology. Fully-supervised models have already achieved outstanding performance with dense pixel-level annotations. However, drawing such labels on the giga-pixel whole slide images is extremely expensive and time-consuming. In this paper, we use only patch-level classification labels to achieve tissue semantic segmentation on histopathology images, finally reducing the annotation efforts. We proposed a two-step model including a classification and a segmentation phases. In the classification phase, we proposed a CAM-based model to generate pseudo masks by patch-level labels. In the segmentation phase, we achieved tissue semantic segmentation by our proposed Multi-Layer Pseudo-Supervision. Several technical novelties have been proposed to reduce the information gap between pixel-level and patch-level annotations. As a part of this paper, we introduced a new weakly-supervised semantic segmentation (WSSS) dataset for lung adenocarcinoma (LUAD-HistoSeg). We conducted several experiments to evaluate our proposed model on two datasets. Our proposed model outperforms two state-of-the-art WSSS approaches. Note that we can achieve comparable quantitative and qualitative results with the fully-supervised model, with only around a 2\% gap for MIoU and FwIoU. By comparing with manual labeling, our model can greatly save the annotation time from hours to minutes. The source code is available at: \url{https://github.com/ChuHan89/WSSS-Tissue}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge