Wuwei Ren
Systematic validation of time-resolved diffuse optical simulators via non-contact SPAD-based measurements
Nov 17, 2025



Abstract:Objective: Time-domain diffuse optical imaging (DOI) requires accurate forward models for photon propagation in scattering media. However, existing simulators lack comprehensive experimental validation, especially for non-contact configurations with oblique illumination. This study rigorously evaluates three widely used open-source simulators, including MMC, NIRFASTer, and Toast++, using time-resolved experimental data. Approach: All simulations employed a unified mesh and point-source illumination. Virtual source correction was applied to FEM solvers for oblique incidence. A time-resolved DOI system with a 32 $\times$ 32 single-photon avalanche diode (SPAD) array acquired transmission-mode data from 16 standardized phantoms with varying absorption coefficient $μ_a$ and reduced scattering coefficient $μ_s'$. The simulation results were quantified across five metrics: spatial-domain (SD) precision, time-domain (TD) precision, oblique beam accuracy, computational speed, and mesh-density independence. Results: Among three simulators, MMC achieves superior accuracy in SD and TD metrics, and shows robustness across all optical properties. NIRFASTer and Toast++ demonstrate comparable overall performance. In general, MMC is optimal for accuracy-critical TD-DOI applications, while NIRFASTer and Toast++ suit scenarios prioritizing speed with sufficiently large $μ_s'$. Besides, virtual source correction is essential for non-contact FEM modeling, which reduced average errors by > 34% in large-angle scenarios. Significance: This work provides benchmarked guidelines for simulator selection during the development phase of next-generation TD-DOI systems. Our work represents the first study to systematically validate TD simulators against SPAD array-based data under clinically relevant non-contact conditions, bridging a critical gap in biomedical optical simulation standards.
$μ$NeuFMT: Optical-Property-Adaptive Fluorescence Molecular Tomography via Implicit Neural Representation
Nov 06, 2025Abstract:Fluorescence Molecular Tomography (FMT) is a promising technique for non-invasive 3D visualization of fluorescent probes, but its reconstruction remains challenging due to the inherent ill-posedness and reliance on inaccurate or often-unknown tissue optical properties. While deep learning methods have shown promise, their supervised nature limits generalization beyond training data. To address these problems, we propose $\mu$NeuFMT, a self-supervised FMT reconstruction framework that integrates implicit neural-based scene representation with explicit physical modeling of photon propagation. Its key innovation lies in jointly optimize both the fluorescence distribution and the optical properties ($\mu$) during reconstruction, eliminating the need for precise prior knowledge of tissue optics or pre-conditioned training data. We demonstrate that $\mu$NeuFMT robustly recovers accurate fluorophore distributions and optical coefficients even with severely erroneous initial values (0.5$\times$ to 2$\times$ of ground truth). Extensive numerical, phantom, and in vivo validations show that $\mu$NeuFMT outperforms conventional and supervised deep learning approaches across diverse heterogeneous scenarios. Our work establishes a new paradigm for robust and accurate FMT reconstruction, paving the way for more reliable molecular imaging in complex clinically related scenarios, such as fluorescence guided surgery.
High-resolution tomographic reconstruction of optical absorbance through scattering media using neural fields
Apr 04, 2023Abstract:Light scattering imposes a major obstacle for imaging objects seated deeply in turbid media, such as biological tissues and foggy air. Diffuse optical tomography (DOT) tackles scattering by volumetrically recovering the optical absorbance and has shown significance in medical imaging, remote sensing and autonomous driving. A conventional DOT reconstruction paradigm necessitates discretizing the object volume into voxels at a pre-determined resolution for modelling diffuse light propagation and the resulting spatial resolution of the reconstruction is generally limited. We propose NeuDOT, a novel DOT scheme based on neural fields (NF) to continuously encode the optical absorbance within the volume and subsequently bridge the gap between model accuracy and high resolution. Comprehensive experiments demonstrate that NeuDOT achieves submillimetre lateral resolution and resolves complex 3D objects at 14 mm-depth, outperforming the state-of-the-art methods. NeuDOT is a non-invasive, high-resolution and computationally efficient tomographic method, and unlocks further applications of NF involving light scattering.
Deep learning facilitates fully automated brain image registration of optoacoustic tomography and magnetic resonance imaging
Sep 04, 2021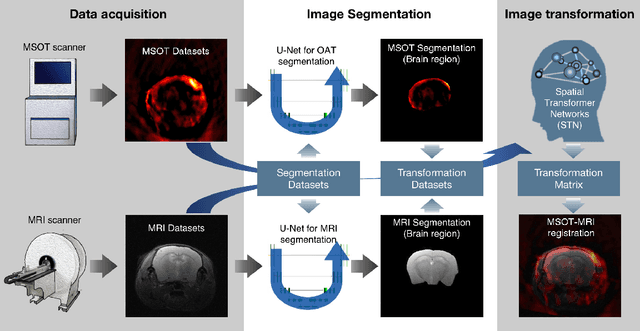
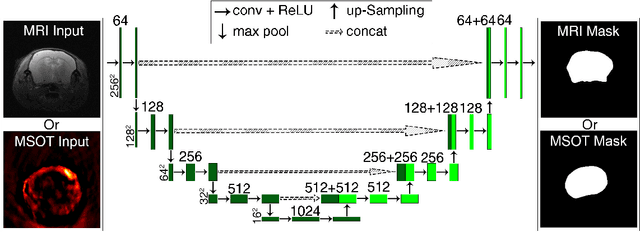
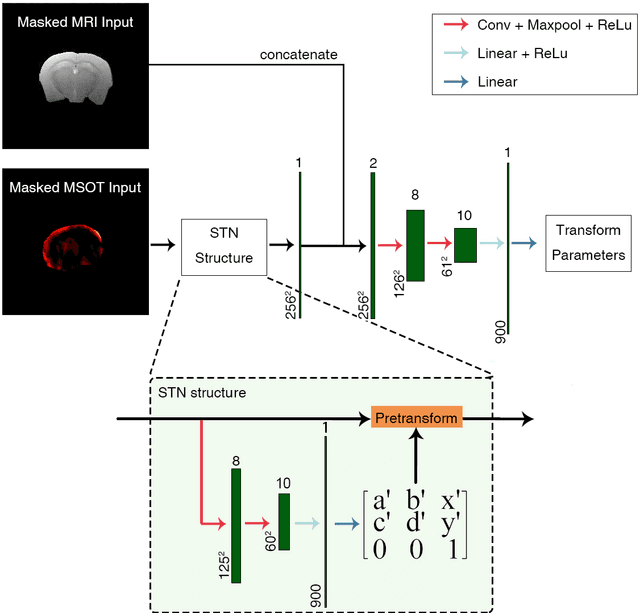
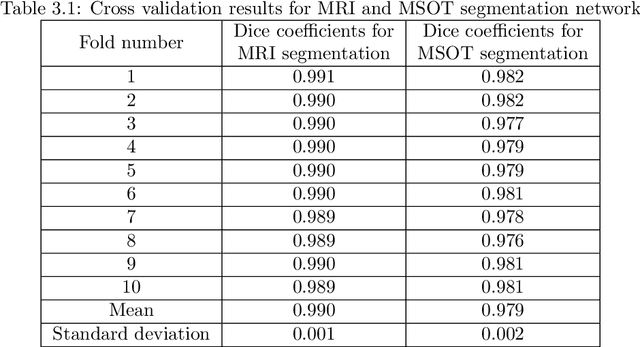
Abstract:Multi-spectral optoacoustic tomography (MSOT) is an emerging optical imaging method providing multiplex molecular and functional information from the rodent brain. It can be greatly augmented by magnetic resonance imaging (MRI) that offers excellent soft-tissue contrast and high-resolution brain anatomy. Nevertheless, registration of multi-modal images remains challenging, chiefly due to the entirely different image contrast rendered by these modalities. Previously reported registration algorithms mostly relied on manual user-dependent brain segmentation, which compromised data interpretation and accurate quantification. Here we propose a fully automated registration method for MSOT-MRI multimodal imaging empowered by deep learning. The automated workflow includes neural network-based image segmentation to generate suitable masks, which are subsequently registered using an additional neural network. Performance of the algorithm is showcased with datasets acquired by cross-sectional MSOT and high-field MRI preclinical scanners. The automated registration method is further validated with manual and half-automated registration, demonstrating its robustness and accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge