Vajira Thambawita
Segmentation Consistency Training: Out-of-Distribution Generalization for Medical Image Segmentation
May 30, 2022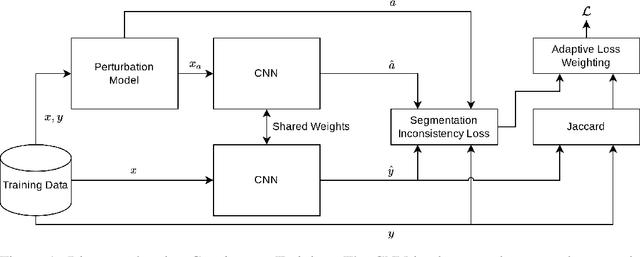

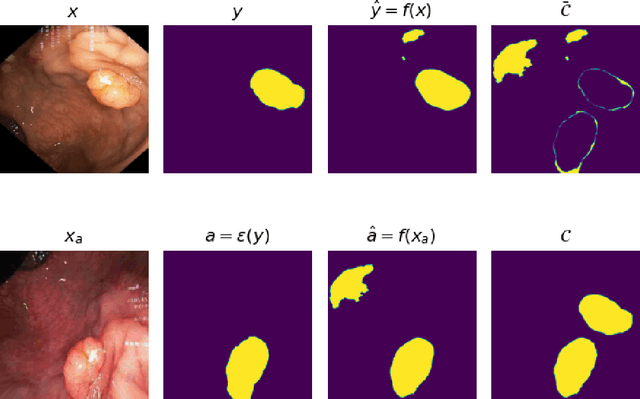
Abstract:Generalizability is seen as one of the major challenges in deep learning, in particular in the domain of medical imaging, where a change of hospital or in imaging routines can lead to a complete failure of a model. To tackle this, we introduce Consistency Training, a training procedure and alternative to data augmentation based on maximizing models' prediction consistency across augmented and unaugmented data in order to facilitate better out-of-distribution generalization. To this end, we develop a novel region-based segmentation loss function called Segmentation Inconsistency Loss (SIL), which considers the differences between pairs of augmented and unaugmented predictions and labels. We demonstrate that Consistency Training outperforms conventional data augmentation on several out-of-distribution datasets on polyp segmentation, a popular medical task.
PolypConnect: Image inpainting for generating realistic gastrointestinal tract images with polyps
May 30, 2022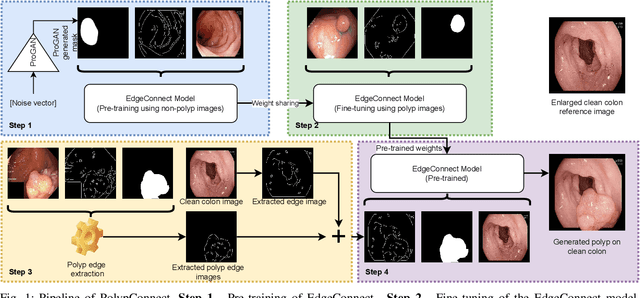
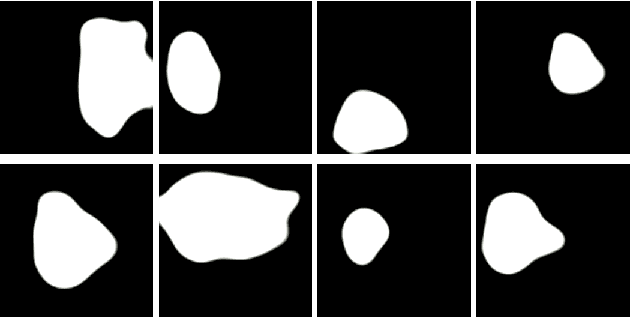

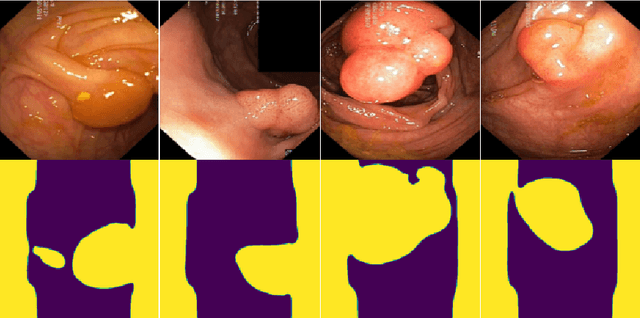
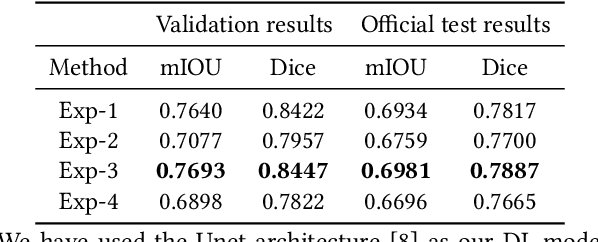
Abstract:Early identification of a polyp in the lower gastrointestinal (GI) tract can lead to prevention of life-threatening colorectal cancer. Developing computer-aided diagnosis (CAD) systems to detect polyps can improve detection accuracy and efficiency and save the time of the domain experts called endoscopists. Lack of annotated data is a common challenge when building CAD systems. Generating synthetic medical data is an active research area to overcome the problem of having relatively few true positive cases in the medical domain. To be able to efficiently train machine learning (ML) models, which are the core of CAD systems, a considerable amount of data should be used. In this respect, we propose the PolypConnect pipeline, which can convert non-polyp images into polyp images to increase the size of training datasets for training. We present the whole pipeline with quantitative and qualitative evaluations involving endoscopists. The polyp segmentation model trained using synthetic data, and real data shows a 5.1% improvement of mean intersection over union (mIOU), compared to the model trained only using real data. The codes of all the experiments are available on GitHub to reproduce the results.
Grid HTM: Hierarchical Temporal Memory for Anomaly Detection in Videos
May 30, 2022
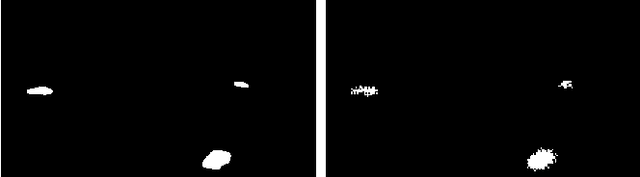
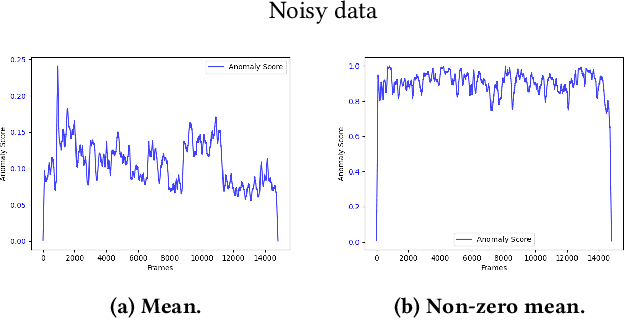
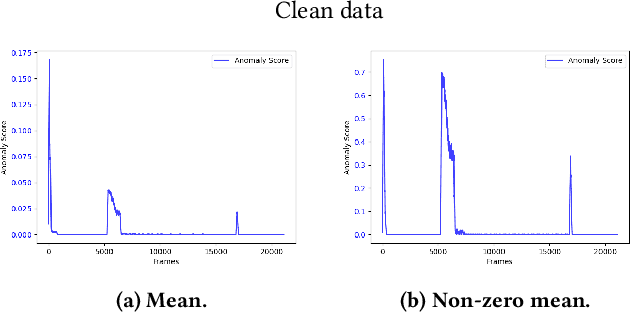
Abstract:The interest for video anomaly detection systems has gained traction for the past few years. The current approaches use deep learning to perform anomaly detection in videos, but this approach has multiple problems. For starters, deep learning in general has issues with noise, concept drift, explainability, and training data volumes. Additionally, anomaly detection in itself is a complex task and faces challenges such as unknowness, heterogeneity, and class imbalance. Anomaly detection using deep learning is therefore mainly constrained to generative models such as generative adversarial networks and autoencoders due to their unsupervised nature, but even they suffer from general deep learning issues and are hard to train properly. In this paper, we explore the capabilities of the Hierarchical Temporal Memory (HTM) algorithm to perform anomaly detection in videos, as it has favorable properties such as noise tolerance and online learning which combats concept drift. We introduce a novel version of HTM, namely, Grid HTM, which is an HTM-based architecture specifically for anomaly detection in complex videos such as surveillance footage.
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022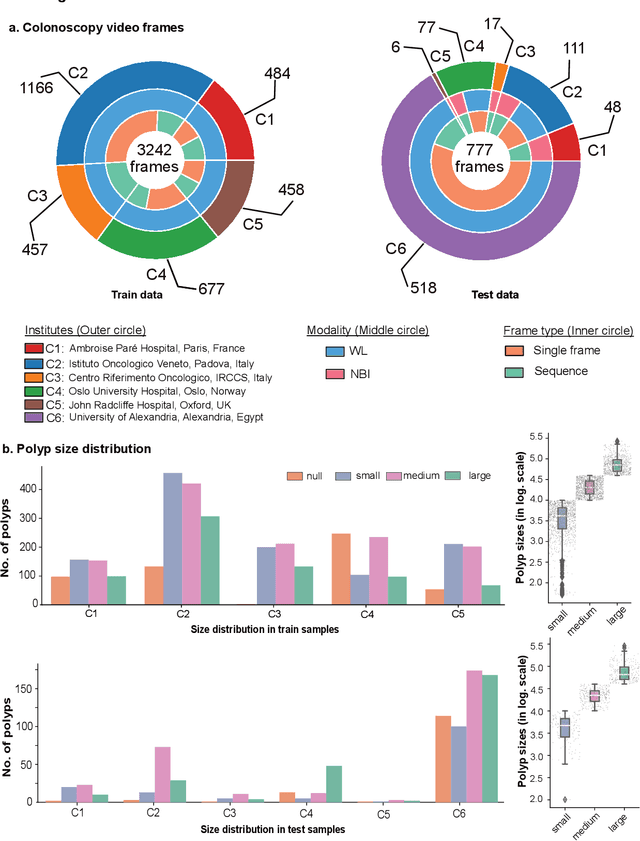
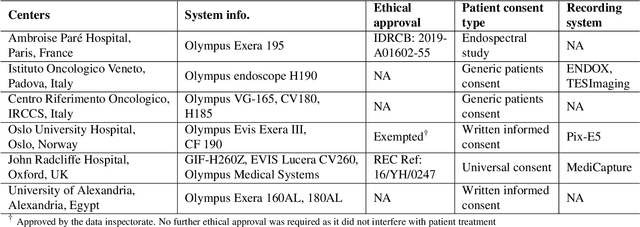
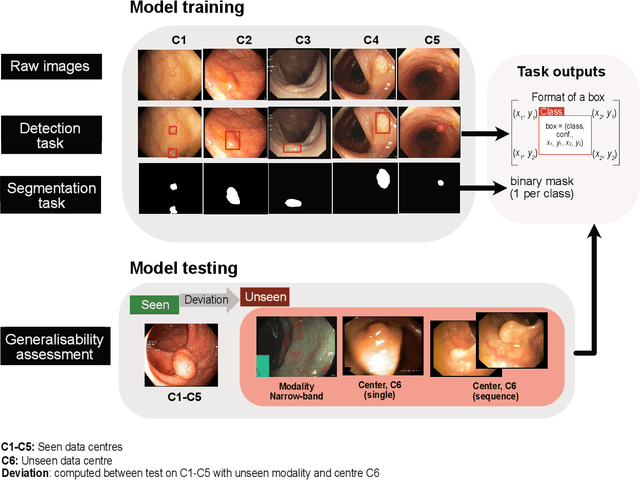
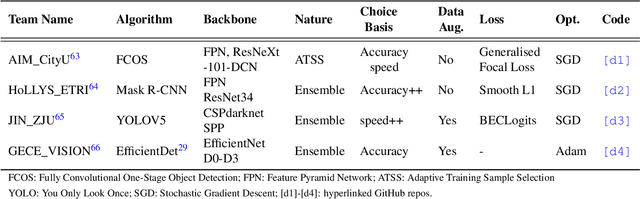
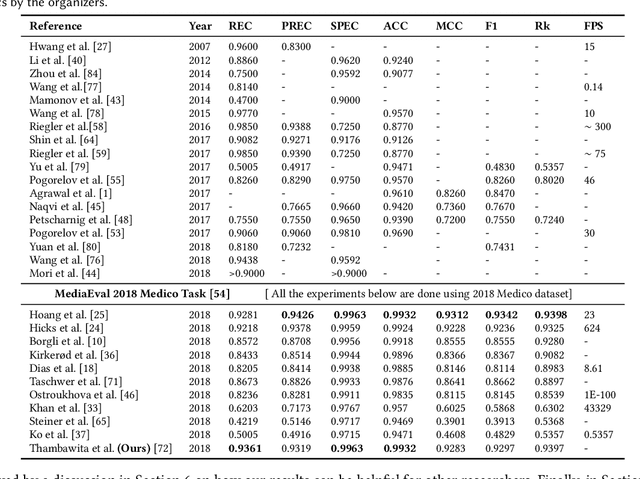
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
MMSys'22 Grand Challenge on AI-based Video Production for Soccer
Feb 02, 2022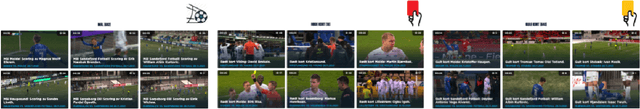
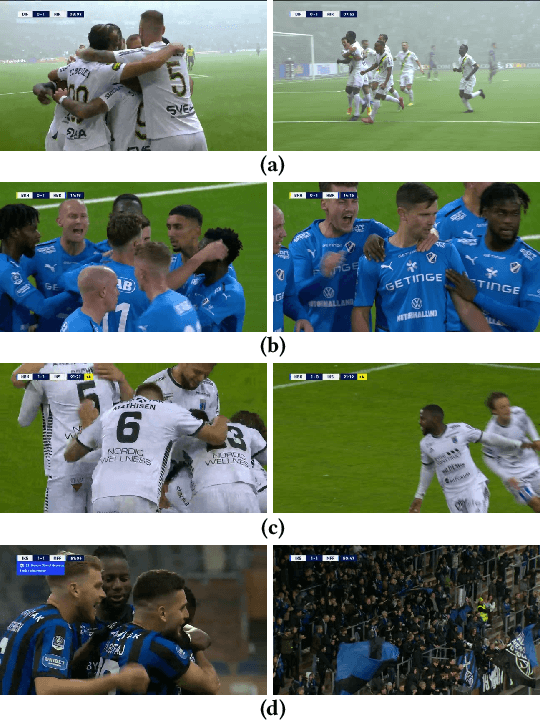
Abstract:Soccer has a considerable market share of the global sports industry, and the interest in viewing videos from soccer games continues to grow. In this respect, it is important to provide game summaries and highlights of the main game events. However, annotating and producing events and summaries often require expensive equipment and a lot of tedious, cumbersome, manual labor. Therefore, automating the video production pipeline providing fast game highlights at a much lower cost is seen as the "holy grail". In this context, recent developments in Artificial Intelligence (AI) technology have shown great potential. Still, state-of-the-art approaches are far from being adequate for practical scenarios that have demanding real-time requirements, as well as strict performance criteria (where at least the detection of official events such as goals and cards must be 100% accurate). In addition, event detection should be thoroughly enhanced by annotation and classification, proper clipping, generating short descriptions, selecting appropriate thumbnails for highlight clips, and finally, combining the event highlights into an overall game summary, similar to what is commonly aired during sports news. Even though the event tagging operation has by far received the most attention, an end-to-end video production pipeline also includes various other operations which serve the overall purpose of automated soccer analysis. This challenge aims to assist the automation of such a production pipeline using AI. In particular, we focus on the enhancement operations that take place after an event has been detected, namely event clipping (Task 1), thumbnail selection (Task 2), and game summarization (Task 3). Challenge website: https://mmsys2022.ie/authors/grand-challenge.
DivergentNets: Medical Image Segmentation by Network Ensemble
Jul 01, 2021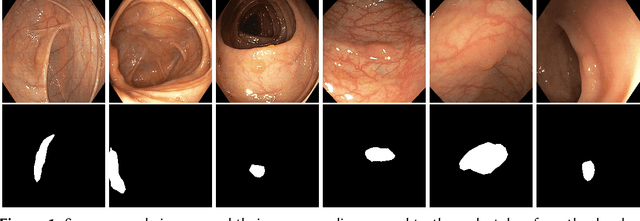
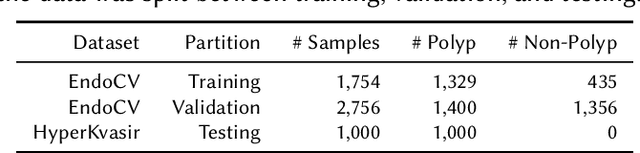
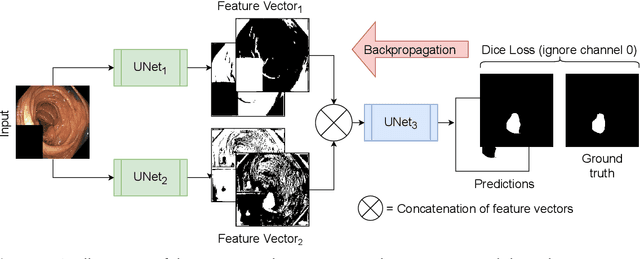
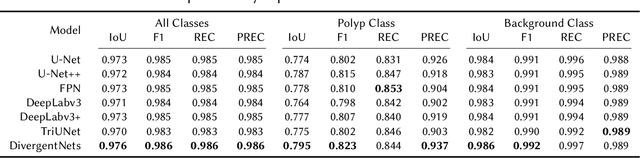
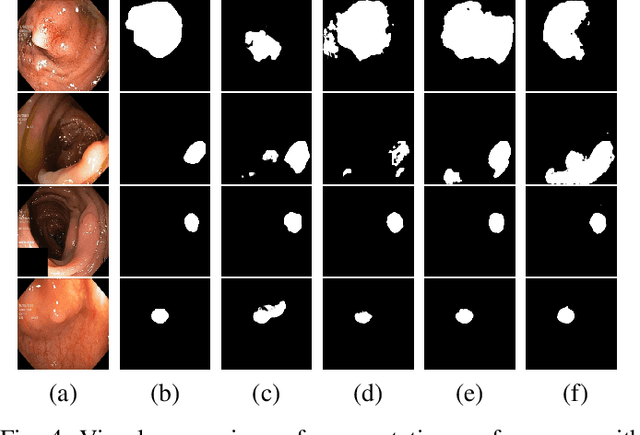
Abstract:Detection of colon polyps has become a trending topic in the intersecting fields of machine learning and gastrointestinal endoscopy. The focus has mainly been on per-frame classification. More recently, polyp segmentation has gained attention in the medical community. Segmentation has the advantage of being more accurate than per-frame classification or object detection as it can show the affected area in greater detail. For our contribution to the EndoCV 2021 segmentation challenge, we propose two separate approaches. First, a segmentation model named TriUNet composed of three separate UNet models. Second, we combine TriUNet with an ensemble of well-known segmentation models, namely UNet++, FPN, DeepLabv3, and DeepLabv3+, into a model called DivergentNets to produce more generalizable medical image segmentation masks. In addition, we propose a modified Dice loss that calculates loss only for a single class when performing multiclass segmentation, forcing the model to focus on what is most important. Overall, the proposed methods achieved the best average scores for each respective round in the challenge, with TriUNet being the winning model in Round I and DivergentNets being the winning model in Round II of the segmentation generalization challenge at EndoCV 2021. The implementation of our approach is made publicly available on GitHub.
* the winning model of the segmentation generalization challenge at EndoCV 2021
SinGAN-Seg: Synthetic Training Data Generation for Medical Image Segmentation
Jun 29, 2021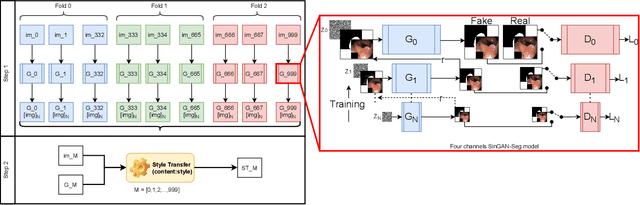
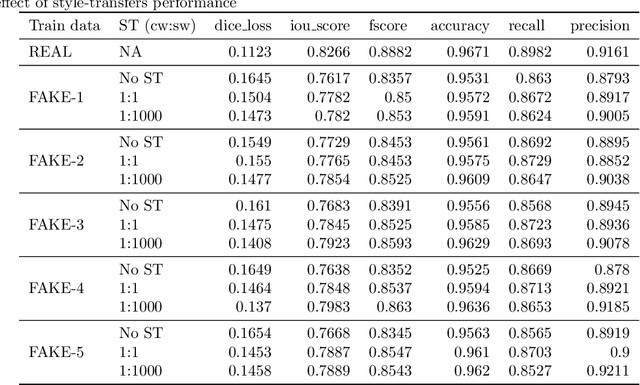
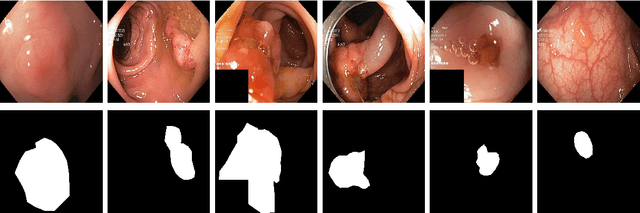
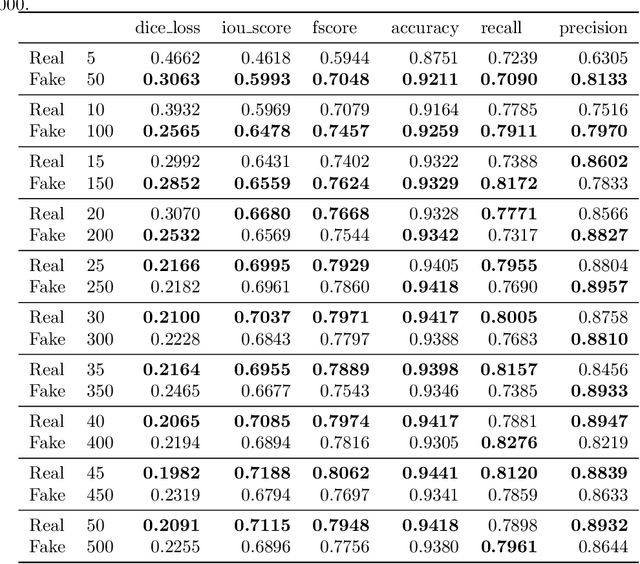
Abstract:Processing medical data to find abnormalities is a time-consuming and costly task, requiring tremendous efforts from medical experts. Therefore, Ai has become a popular tool for the automatic processing of medical data, acting as a supportive tool for doctors. AI tools highly depend on data for training the models. However, there are several constraints to access to large amounts of medical data to train machine learning algorithms in the medical domain, e.g., due to privacy concerns and the costly, time-consuming medical data annotation process. To address this, in this paper we present a novel synthetic data generation pipeline called SinGAN-Seg to produce synthetic medical data with the corresponding annotated ground truth masks. We show that these synthetic data generation pipelines can be used as an alternative to bypass privacy concerns and as an alternative way to produce artificial segmentation datasets with corresponding ground truth masks to avoid the tedious medical data annotation process. As a proof of concept, we used an open polyp segmentation dataset. By training UNet++ using both the real polyp segmentation dataset and the corresponding synthetic dataset generated from the SinGAN-Seg pipeline, we show that the synthetic data can achieve a very close performance to the real data when the real segmentation datasets are large enough. In addition, we show that synthetic data generated from the SinGAN-Seg pipeline improving the performance of segmentation algorithms when the training dataset is very small. Since our SinGAN-Seg pipeline is applicable for any medical dataset, this pipeline can be used with any other segmentation datasets.
Few-shot segmentation of medical images based on meta-learning with implicit gradients
Jun 06, 2021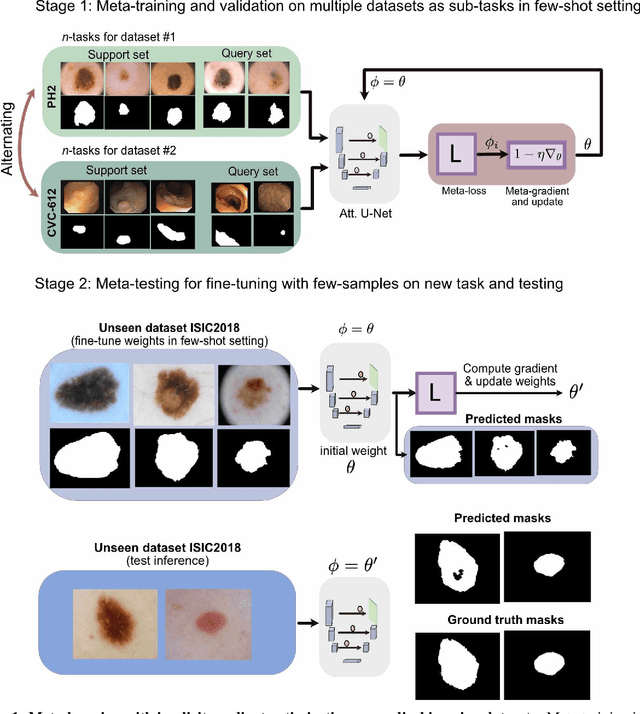
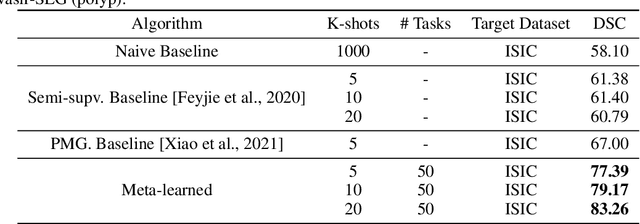
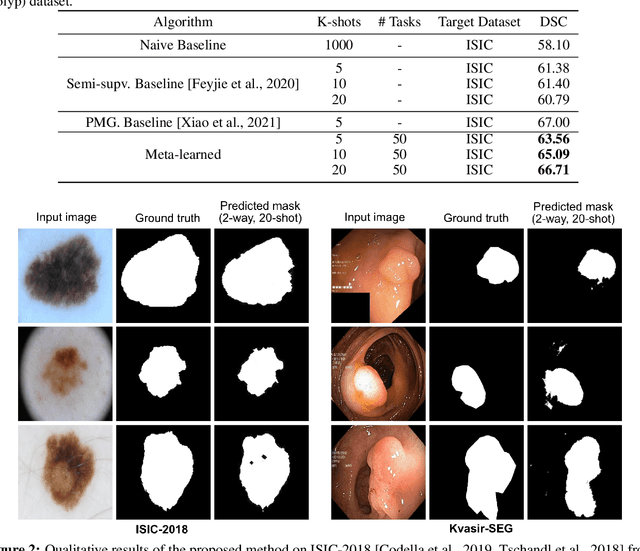
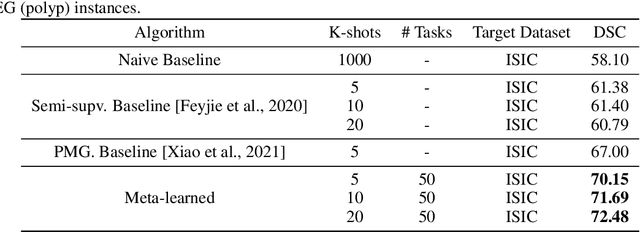
Abstract:Classical supervised methods commonly used often suffer from the requirement of an abudant number of training samples and are unable to generalize on unseen datasets. As a result, the broader application of any trained model is very limited in clinical settings. However, few-shot approaches can minimize the need for enormous reliable ground truth labels that are both labor intensive and expensive. To this end, we propose to exploit an optimization-based implicit model agnostic meta-learning {iMAML} algorithm in a few-shot setting for medical image segmentation. Our approach can leverage the learned weights from a diverse set of training samples and can be deployed on a new unseen dataset. We show that unlike classical few-shot learning approaches, our method has improved generalization capability. To our knowledge, this is the first work that exploits iMAML for medical image segmentation. Our quantitative results on publicly available skin and polyp datasets show that the proposed method outperforms the naive supervised baseline model and two recent few-shot segmentation approaches by large margins.
Pyramid-Focus-Augmentation: Medical Image Segmentation with Step-Wise Focus
Dec 14, 2020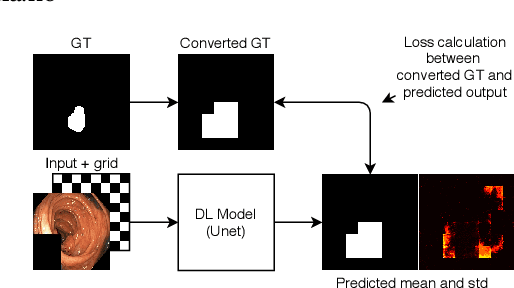
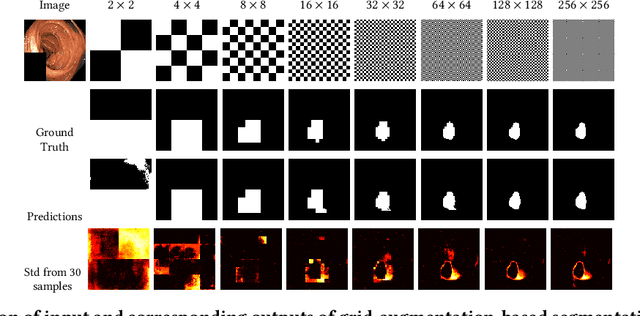
Abstract:Segmentation of findings in the gastrointestinal tract is a challenging but also an important task which is an important building stone for sufficient automatic decision support systems. In this work, we present our solution for the Medico 2020 task, which focused on the problem of colon polyp segmentation. We present our simple but efficient idea of using an augmentation method that uses grids in a pyramid-like manner (large to small) for segmentation. Our results show that the proposed methods work as indented and can also lead to comparable results when competing with other methods.
An Extensive Study on Cross-Dataset Bias and Evaluation Metrics Interpretation for Machine Learning applied to Gastrointestinal Tract Abnormality Classification
May 08, 2020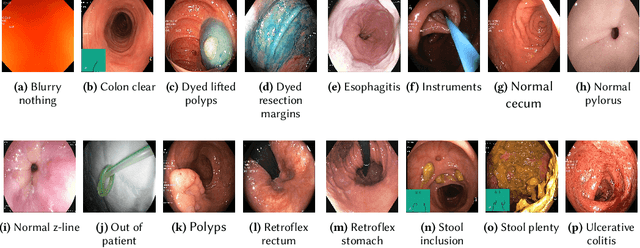
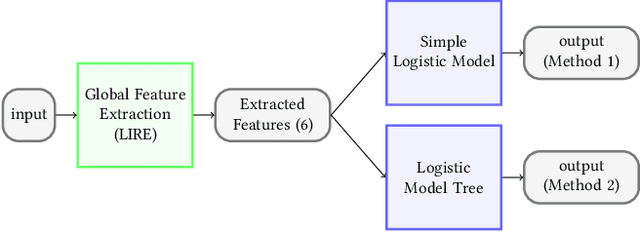
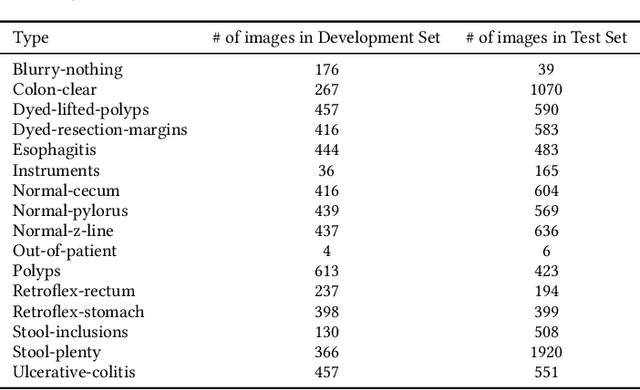
Abstract:Precise and efficient automated identification of Gastrointestinal (GI) tract diseases can help doctors treat more patients and improve the rate of disease detection and identification. Currently, automatic analysis of diseases in the GI tract is a hot topic in both computer science and medical-related journals. Nevertheless, the evaluation of such an automatic analysis is often incomplete or simply wrong. Algorithms are often only tested on small and biased datasets, and cross-dataset evaluations are rarely performed. A clear understanding of evaluation metrics and machine learning models with cross datasets is crucial to bring research in the field to a new quality level. Towards this goal, we present comprehensive evaluations of five distinct machine learning models using Global Features and Deep Neural Networks that can classify 16 different key types of GI tract conditions, including pathological findings, anatomical landmarks, polyp removal conditions, and normal findings from images captured by common GI tract examination instruments. In our evaluation, we introduce performance hexagons using six performance metrics such as recall, precision, specificity, accuracy, F1-score, and Matthews Correlation Coefficient to demonstrate how to determine the real capabilities of models rather than evaluating them shallowly. Furthermore, we perform cross-dataset evaluations using different datasets for training and testing. With these cross-dataset evaluations, we demonstrate the challenge of actually building a generalizable model that could be used across different hospitals. Our experiments clearly show that more sophisticated performance metrics and evaluation methods need to be applied to get reliable models rather than depending on evaluations of the splits of the same dataset, i.e., the performance metrics should always be interpreted together rather than relying on a single metric.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge