Shangyang Li
Agri-R1: Empowering Generalizable Agricultural Reasoning in Vision-Language Models with Reinforcement Learning
Jan 08, 2026Abstract:Agricultural disease diagnosis challenges VLMs, as conventional fine-tuning requires extensive labels, lacks interpretability, and generalizes poorly. While reasoning improves model robustness, existing methods rely on costly expert annotations and rarely address the open-ended, diverse nature of agricultural queries. To address these limitations, we propose \textbf{Agri-R1}, a reasoning-enhanced large model for agriculture. Our framework automates high-quality reasoning data generation via vision-language synthesis and LLM-based filtering, using only 19\% of available samples. Training employs Group Relative Policy Optimization (GRPO) with a novel proposed reward function that integrates domain-specific lexicons and fuzzy matching to assess both correctness and linguistic flexibility in open-ended responses. Evaluated on CDDMBench, our resulting 3B-parameter model achieves performance competitive with 7B- to 13B-parameter baselines, showing a +23.2\% relative gain in disease recognition accuracy, +33.3\% in agricultural knowledge QA, and a +26.10-point improvement in cross-domain generalization over standard fine-tuning. Ablation studies confirm that the synergy between structured reasoning data and GRPO-driven exploration underpins these gains, with benefits scaling as question complexity increases.
Beyond Classification Accuracy: Neural-MedBench and the Need for Deeper Reasoning Benchmarks
Sep 26, 2025Abstract:Recent advances in vision-language models (VLMs) have achieved remarkable performance on standard medical benchmarks, yet their true clinical reasoning ability remains unclear. Existing datasets predominantly emphasize classification accuracy, creating an evaluation illusion in which models appear proficient while still failing at high-stakes diagnostic reasoning. We introduce Neural-MedBench, a compact yet reasoning-intensive benchmark specifically designed to probe the limits of multimodal clinical reasoning in neurology. Neural-MedBench integrates multi-sequence MRI scans, structured electronic health records, and clinical notes, and encompasses three core task families: differential diagnosis, lesion recognition, and rationale generation. To ensure reliable evaluation, we develop a hybrid scoring pipeline that combines LLM-based graders, clinician validation, and semantic similarity metrics. Through systematic evaluation of state-of-the-art VLMs, including GPT-4o, Claude-4, and MedGemma, we observe a sharp performance drop compared to conventional datasets. Error analysis shows that reasoning failures, rather than perceptual errors, dominate model shortcomings. Our findings highlight the necessity of a Two-Axis Evaluation Framework: breadth-oriented large datasets for statistical generalization, and depth-oriented, compact benchmarks such as Neural-MedBench for reasoning fidelity. We release Neural-MedBench at https://neuromedbench.github.io/ as an open and extensible diagnostic testbed, which guides the expansion of future benchmarks and enables rigorous yet cost-effective assessment of clinically trustworthy AI.
A differentiable brain simulator bridging brain simulation and brain-inspired computing
Nov 09, 2023



Abstract:Brain simulation builds dynamical models to mimic the structure and functions of the brain, while brain-inspired computing (BIC) develops intelligent systems by learning from the structure and functions of the brain. The two fields are intertwined and should share a common programming framework to facilitate each other's development. However, none of the existing software in the fields can achieve this goal, because traditional brain simulators lack differentiability for training, while existing deep learning (DL) frameworks fail to capture the biophysical realism and complexity of brain dynamics. In this paper, we introduce BrainPy, a differentiable brain simulator developed using JAX and XLA, with the aim of bridging the gap between brain simulation and BIC. BrainPy expands upon the functionalities of JAX, a powerful AI framework, by introducing complete capabilities for flexible, efficient, and scalable brain simulation. It offers a range of sparse and event-driven operators for efficient and scalable brain simulation, an abstraction for managing the intricacies of synaptic computations, a modular and flexible interface for constructing multi-scale brain models, and an object-oriented just-in-time compilation approach to handle the memory-intensive nature of brain dynamics. We showcase the efficiency and scalability of BrainPy on benchmark tasks, highlight its differentiable simulation for biologically plausible spiking models, and discuss its potential to support research at the intersection of brain simulation and BIC.
Learning Robust Representation through Graph Adversarial Contrastive Learning
Jan 31, 2022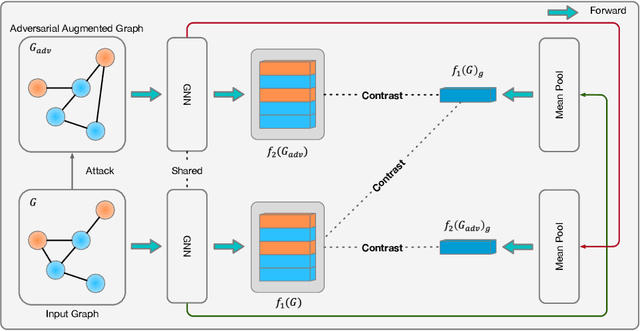
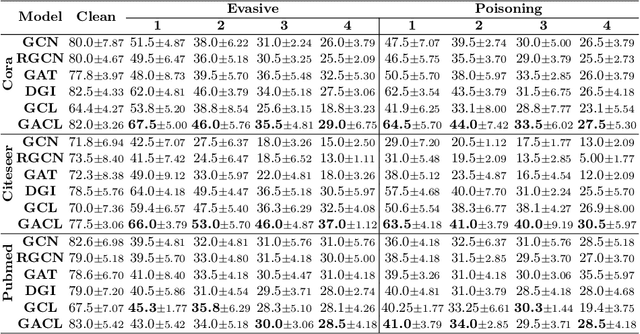
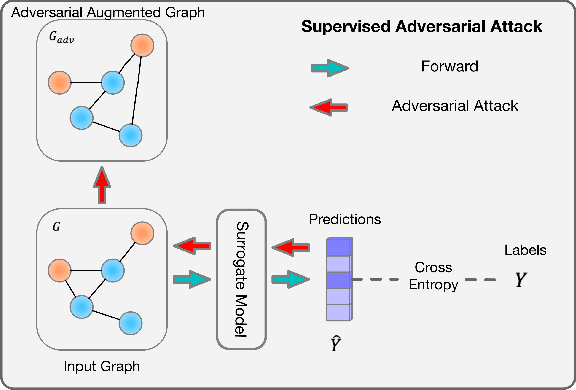
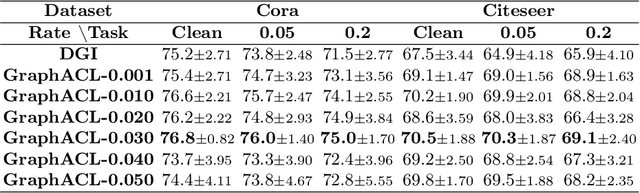
Abstract:Existing studies show that node representations generated by graph neural networks (GNNs) are vulnerable to adversarial attacks, such as unnoticeable perturbations of adjacent matrix and node features. Thus, it is requisite to learn robust representations in graph neural networks. To improve the robustness of graph representation learning, we propose a novel Graph Adversarial Contrastive Learning framework (GraphACL) by introducing adversarial augmentations into graph self-supervised learning. In this framework, we maximize the mutual information between local and global representations of a perturbed graph and its adversarial augmentations, where the adversarial graphs can be generated in either supervised or unsupervised approaches. Based on the Information Bottleneck Principle, we theoretically prove that our method could obtain a much tighter bound, thus improving the robustness of graph representation learning. Empirically, we evaluate several methods on a range of node classification benchmarks and the results demonstrate GraphACL could achieve comparable accuracy over previous supervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge