Multi-modal Graph Fusion for Inductive Disease Classification in Incomplete Datasets
May 08, 2019Gerome Vivar, Hendrik Burwinkel, Anees Kazi, Andreas Zwergal, Nassir Navab, Seyed-Ahmad Ahmadi
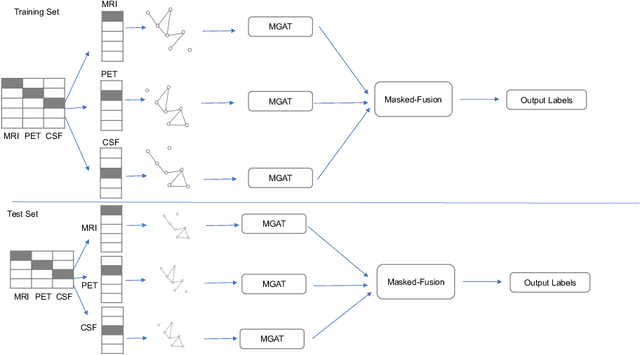
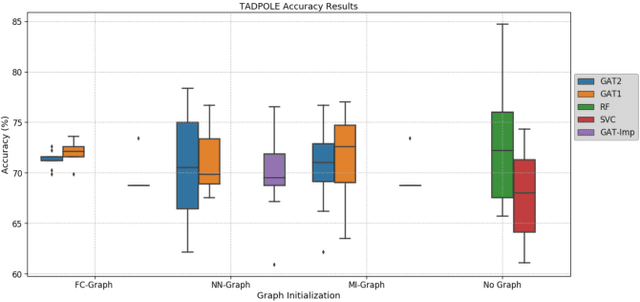
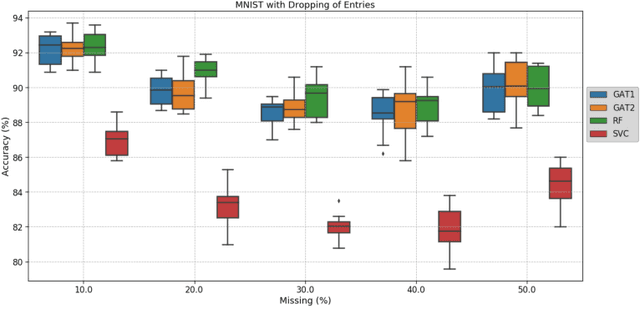
Clinical diagnostic decision making and population-based studies often rely on multi-modal data which is noisy and incomplete. Recently, several works proposed geometric deep learning approaches to solve disease classification, by modeling patients as nodes in a graph, along with graph signal processing of multi-modal features. Many of these approaches are limited by assuming modality- and feature-completeness, and by transductive inference, which requires re-training of the entire model for each new test sample. In this work, we propose a novel inductive graph-based approach that can generalize to out-of-sample patients, despite missing features from entire modalities per patient. We propose multi-modal graph fusion which is trained end-to-end towards node-level classification. We demonstrate the fundamental working principle of this method on a simplified MNIST toy dataset. In experiments on medical data, our method outperforms single static graph approach in multi-modal disease classification.
Adaptive image-feature learning for disease classification using inductive graph networks
May 08, 2019Hendrik Burwinkel, Anees Kazi, Gerome Vivar, Shadi Albarqouni, Guillaume Zahnd, Nassir Navab, Seyed-Ahmad Ahmadi
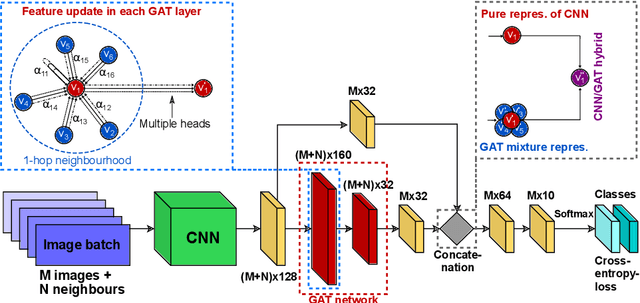
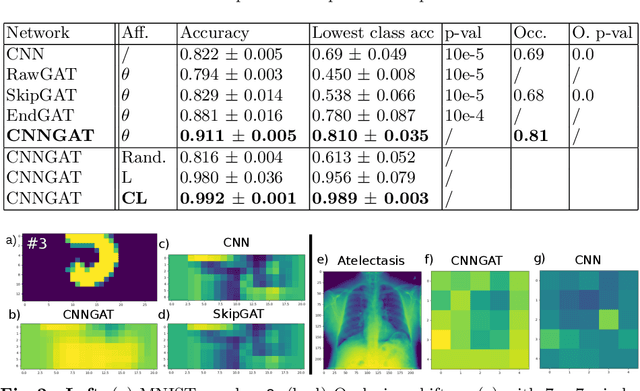
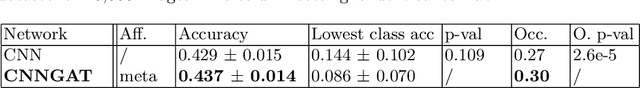
Recently, Geometric Deep Learning (GDL) has been introduced as a novel and versatile framework for computer-aided disease classification. GDL uses patient meta-information such as age and gender to model patient cohort relations in a graph structure. Concepts from graph signal processing are leveraged to learn the optimal mapping of multi-modal features, e.g. from images to disease classes. Related studies so far have considered image features that are extracted in a pre-processing step. We hypothesize that such an approach prevents the network from optimizing feature representations towards achieving the best performance in the graph network. We propose a new network architecture that exploits an inductive end-to-end learning approach for disease classification, where filters from both the CNN and the graph are trained jointly. We validate this architecture against state-of-the-art inductive graph networks and demonstrate significantly improved classification scores on a modified MNIST toy dataset, as well as comparable classification results with higher stability on a chest X-ray image dataset. Additionally, we explain how the structural information of the graph affects both the image filters and the feature learning.
InceptionGCN: Receptive Field Aware Graph Convolutional Network for Disease Prediction
Mar 11, 2019Anees Kazi, Shayan shekarforoush, S. Arvind krishna, Hendrik Burwinkel, Gerome Vivar, Karsten Kortuem, Seyed-Ahmad Ahmadi, Shadi Albarqouni, Nassir Navab
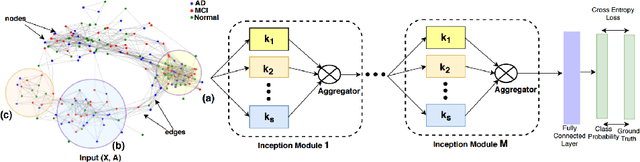
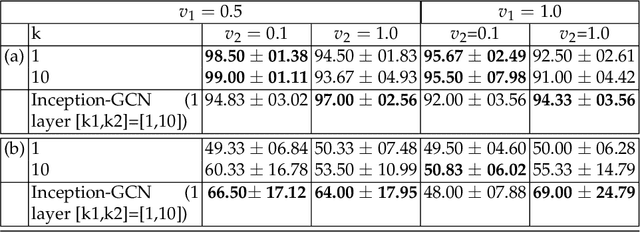
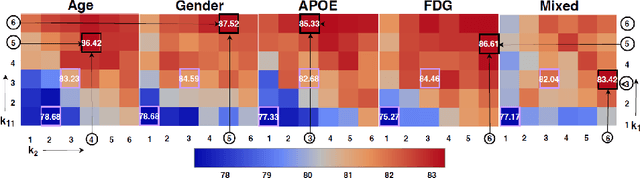
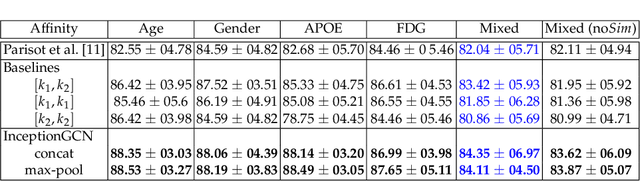
Geometric deep learning provides a principled and versatile manner for the integration of imaging and non-imaging modalities in the medical domain. Graph Convolutional Networks (GCNs) in particular have been explored on a wide variety of problems such as disease prediction, segmentation, and matrix completion by leveraging large, multimodal datasets. In this paper, we introduce a new spectral domain architecture for deep learning on graphs for disease prediction. The novelty lies in defining geometric 'inception modules' which are capable of capturing intra- and inter-graph structural heterogeneity during convolutions. We design filters with different kernel sizes to build our architecture. We show our disease prediction results on two publicly available datasets. Further, we provide insights on the behaviour of regular GCNs and our proposed model under varying input scenarios on simulated data.
Stabilizing Inputs to Approximated Nonlinear Functions for Inference with Homomorphic Encryption in Deep Neural Networks
Feb 05, 2019Moustafa AboulAtta, Matthias Ossadnik, Seyed-Ahmad Ahmadi
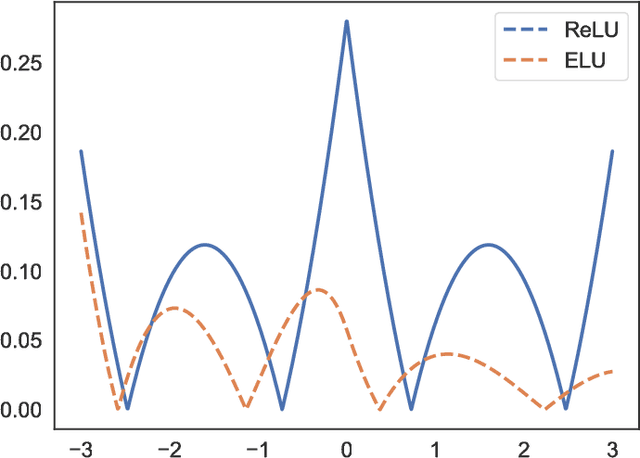
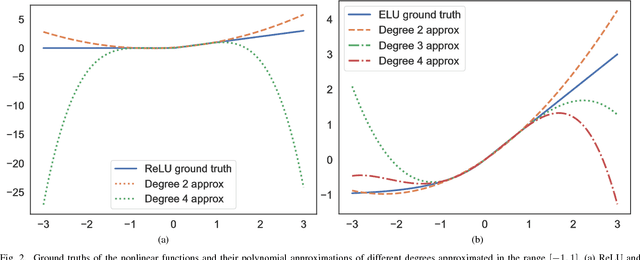
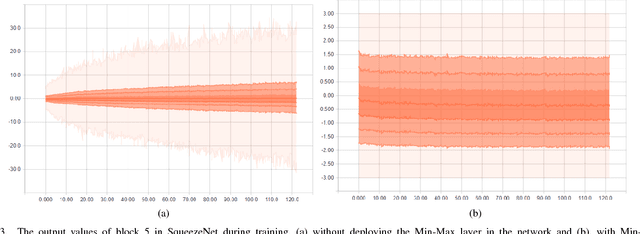
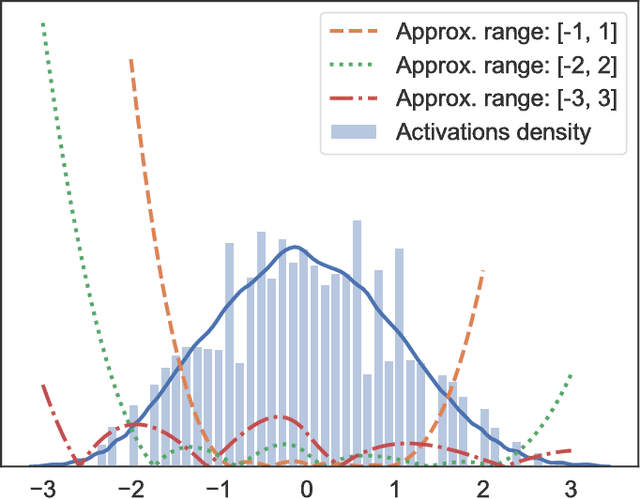
Leveled Homomorphic Encryption (LHE) offers a potential solution that could allow sectors with sensitive data to utilize the cloud and securely deploy their models for remote inference with Deep Neural Networks (DNN). However, this application faces several obstacles due to the limitations of LHE. One of the main problems is the incompatibility of commonly used nonlinear functions in DNN with the operations supported by LHE, i.e. addition and multiplication. As common in LHE approaches, we train a model with a nonlinear function, and replace it with a low-degree polynomial approximation at inference time on private data. While this typically leads to approximation errors and loss in prediction accuracy, we propose a method that reduces this loss to small values or eliminates it entirely, depending on simple hyper-parameters. This is achieved by the introduction of a novel and elegantly simple Min-Max normalization scheme, which scales inputs to nonlinear functions into ranges with low approximation error. While being intuitive in its concept and trivial to implement, we empirically show that it offers a stable and effective approximation solution to nonlinear functions in DNN. In return, this can enable deeper networks with LHE, and facilitate the development of security- and privacy-aware analytics applications.
TOMAAT: volumetric medical image analysis as a cloud service
Apr 25, 2018Fausto Milletari, Johann Frei, Seyed-Ahmad Ahmadi
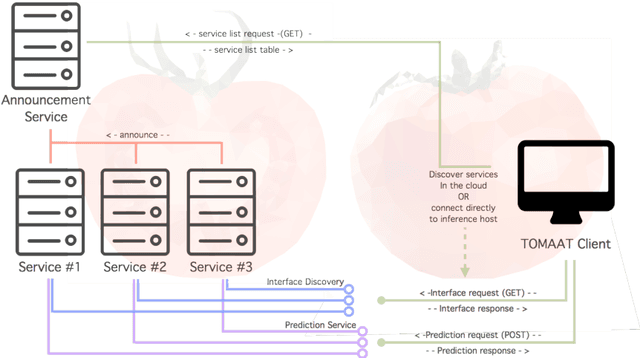
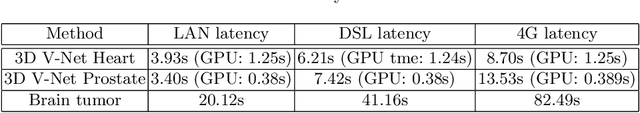
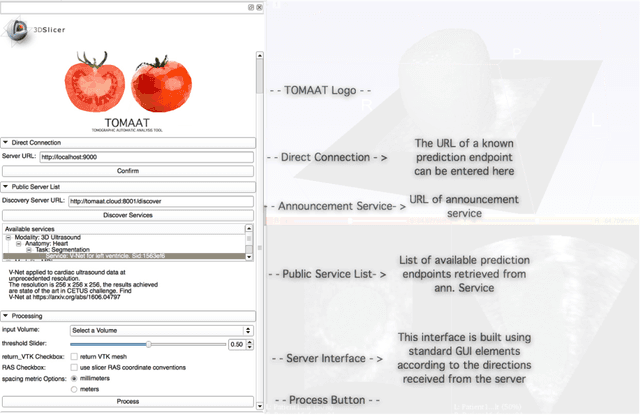
Deep learning has been recently applied to a multitude of computer vision and medical image analysis problems. Although recent research efforts have improved the state of the art, most of the methods cannot be easily accessed, compared or used by either researchers or the general public. Researchers often publish their code and trained models on the internet, but this does not always enable these approaches to be easily used or integrated in stand-alone applications and existing workflows. In this paper we propose a framework which allows easy deployment and access of deep learning methods for segmentation through a cloud-based architecture. Our approach comprises three parts: a server, which wraps trained deep learning models and their pre- and post-processing data pipelines and makes them available on the cloud; a client which interfaces with the server to obtain predictions on user data; a service registry that informs clients about available prediction endpoints that are available in the cloud. These three parts constitute the open-source TOMAAT framework.
Multi-modal Disease Classification in Incomplete Datasets Using Geometric Matrix Completion
Mar 30, 2018Gerome Vivar, Andreas Zwergal, Nassir Navab, Seyed-Ahmad Ahmadi
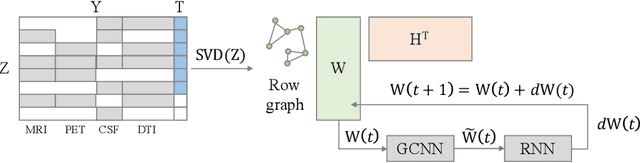
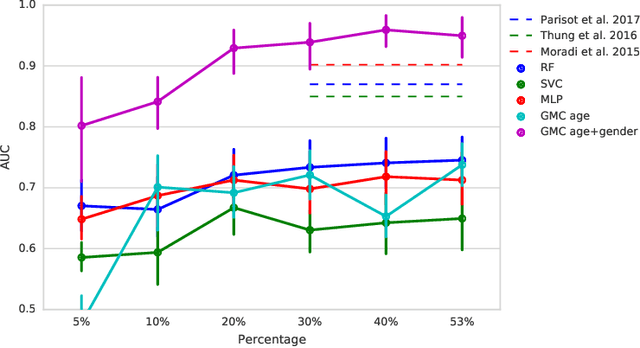
In large population-based studies and in clinical routine, tasks like disease diagnosis and progression prediction are inherently based on a rich set of multi-modal data, including imaging and other sensor data, clinical scores, phenotypes, labels and demographics. However, missing features, rater bias and inaccurate measurements are typical ailments of real-life medical datasets. Recently, it has been shown that deep learning with graph convolution neural networks (GCN) can outperform traditional machine learning in disease classification, but missing features remain an open problem. In this work, we follow up on the idea of modeling multi-modal disease classification as a matrix completion problem, with simultaneous classification and non-linear imputation of features. Compared to methods before, we arrange subjects in a graph-structure and solve classification through geometric matrix completion, which simulates a heat diffusion process that is learned and solved with a recurrent neural network. We demonstrate the potential of this method on the ADNI-based TADPOLE dataset and on the task of predicting the transition from MCI to Alzheimer's disease. With an AUC of 0.950 and classification accuracy of 87%, our approach outperforms standard linear and non-linear classifiers, as well as several state-of-the-art results in related literature, including a recently proposed GCN-based approach.
Classification of sparsely labeled spatio-temporal data through semi-supervised adversarial learning
Jan 29, 2018Atanas Mirchev, Seyed-Ahmad Ahmadi
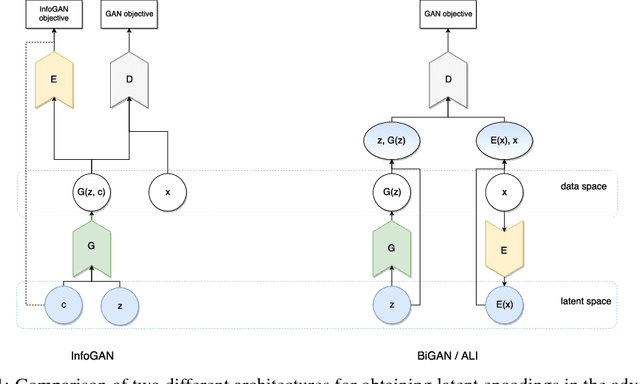
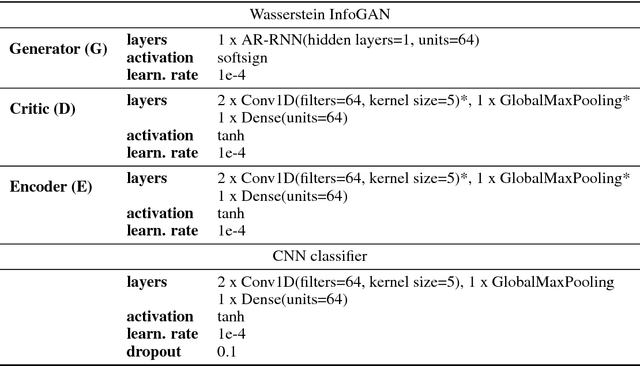
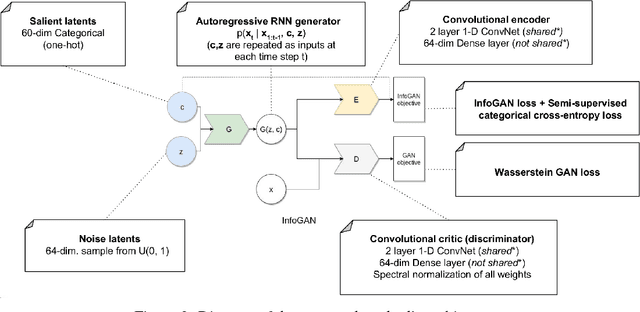
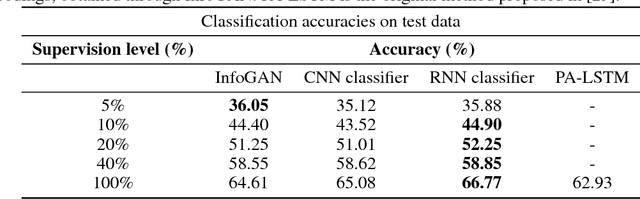
In recent years, Generative Adversarial Networks (GAN) have emerged as a powerful method for learning the mapping from noisy latent spaces to realistic data samples in high-dimensional space. So far, the development and application of GANs have been predominantly focused on spatial data such as images. In this project, we aim at modeling of spatio-temporal sensor data instead, i.e. dynamic data over time. The main goal is to encode temporal data into a global and low-dimensional latent vector that captures the dynamics of the spatio-temporal signal. To this end, we incorporate auto-regressive RNNs, Wasserstein GAN loss, spectral norm weight constraints and a semi-supervised learning scheme into InfoGAN, a method for retrieval of meaningful latents in adversarial learning. To demonstrate the modeling capability of our method, we encode full-body skeletal human motion from a large dataset representing 60 classes of daily activities, recorded in a multi-Kinect setup. Initial results indicate competitive classification performance of the learned latent representations, compared to direct CNN/RNN inference. In future work, we plan to apply this method on a related problem in the medical domain, i.e. on recovery of meaningful latents in gait analysis of patients with vertigo and balance disorders.
Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks
Feb 23, 2017Patrick Ferdinand Christ, Florian Ettlinger, Felix Grün, Mohamed Ezzeldin A. Elshaera, Jana Lipkova, Sebastian Schlecht, Freba Ahmaddy, Sunil Tatavarty, Marc Bickel, Patrick Bilic, Markus Rempfler, Felix Hofmann, Melvin D Anastasi, Seyed-Ahmad Ahmadi, Georgios Kaissis, Julian Holch, Wieland Sommer, Rickmer Braren, Volker Heinemann, Bjoern Menze
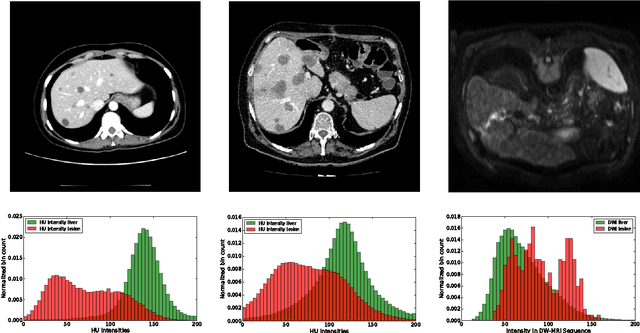
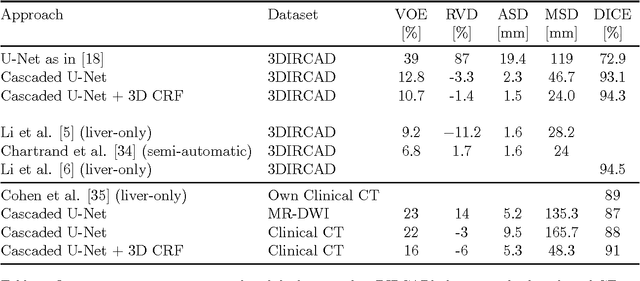
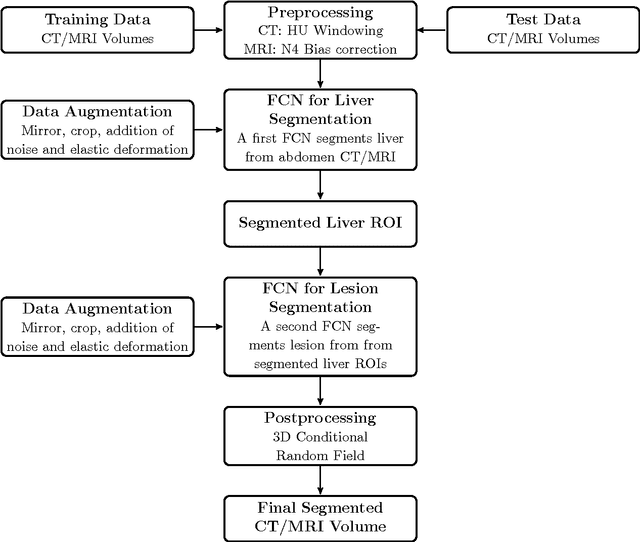
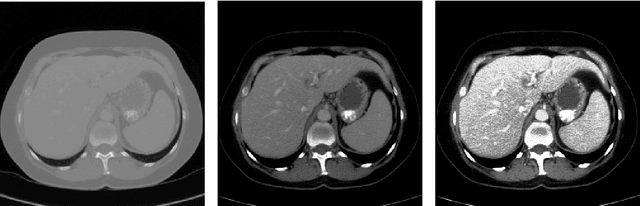
Automatic segmentation of the liver and hepatic lesions is an important step towards deriving quantitative biomarkers for accurate clinical diagnosis and computer-aided decision support systems. This paper presents a method to automatically segment liver and lesions in CT and MRI abdomen images using cascaded fully convolutional neural networks (CFCNs) enabling the segmentation of a large-scale medical trial or quantitative image analysis. We train and cascade two FCNs for a combined segmentation of the liver and its lesions. In the first step, we train a FCN to segment the liver as ROI input for a second FCN. The second FCN solely segments lesions within the predicted liver ROIs of step 1. CFCN models were trained on an abdominal CT dataset comprising 100 hepatic tumor volumes. Validations on further datasets show that CFCN-based semantic liver and lesion segmentation achieves Dice scores over 94% for liver with computation times below 100s per volume. We further experimentally demonstrate the robustness of the proposed method on an 38 MRI liver tumor volumes and the public 3DIRCAD dataset.
SurvivalNet: Predicting patient survival from diffusion weighted magnetic resonance images using cascaded fully convolutional and 3D convolutional neural networks
Feb 20, 2017Patrick Ferdinand Christ, Florian Ettlinger, Georgios Kaissis, Sebastian Schlecht, Freba Ahmaddy, Felix Grün, Alexander Valentinitsch, Seyed-Ahmad Ahmadi, Rickmer Braren, Bjoern Menze
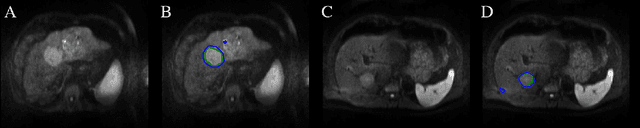

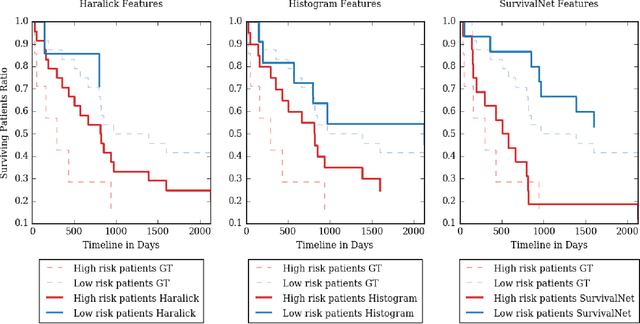
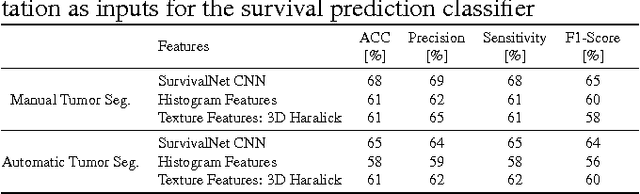
Automatic non-invasive assessment of hepatocellular carcinoma (HCC) malignancy has the potential to substantially enhance tumor treatment strategies for HCC patients. In this work we present a novel framework to automatically characterize the malignancy of HCC lesions from DWI images. We predict HCC malignancy in two steps: As a first step we automatically segment HCC tumor lesions using cascaded fully convolutional neural networks (CFCN). A 3D neural network (SurvivalNet) then predicts the HCC lesions' malignancy from the HCC tumor segmentation. We formulate this task as a classification problem with classes being "low risk" and "high risk" represented by longer or shorter survival times than the median survival. We evaluated our method on DWI of 31 HCC patients. Our proposed framework achieves an end-to-end accuracy of 65% with a Dice score for the automatic lesion segmentation of 69% and an accuracy of 68% for tumor malignancy classification based on expert annotations. We compared the SurvivalNet to classical handcrafted features such as Histogram and Haralick and show experimentally that SurvivalNet outperforms the handcrafted features in HCC malignancy classification. End-to-end assessment of tumor malignancy based on our proposed fully automatic framework corresponds to assessment based on expert annotations with high significance (p>0.95).
Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields
Oct 07, 2016Patrick Ferdinand Christ, Mohamed Ezzeldin A. Elshaer, Florian Ettlinger, Sunil Tatavarty, Marc Bickel, Patrick Bilic, Markus Rempfler, Marco Armbruster, Felix Hofmann, Melvin D'Anastasi, Wieland H. Sommer, Seyed-Ahmad Ahmadi, Bjoern H. Menze
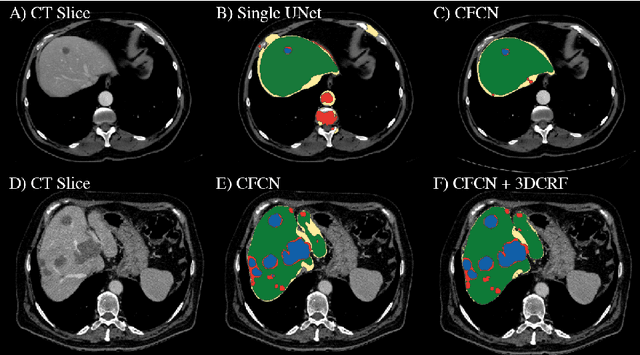
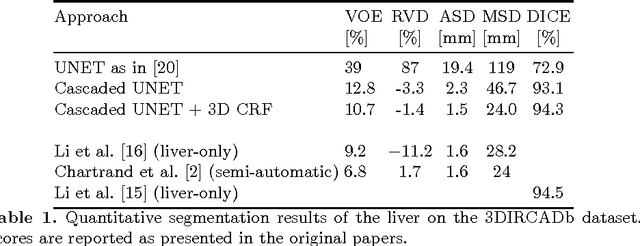
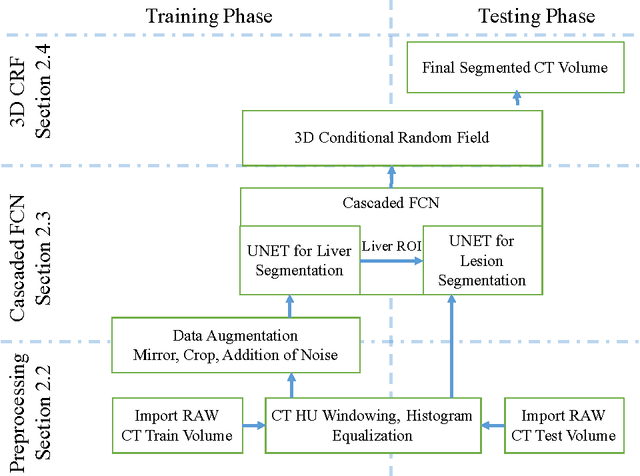
Automatic segmentation of the liver and its lesion is an important step towards deriving quantitative biomarkers for accurate clinical diagnosis and computer-aided decision support systems. This paper presents a method to automatically segment liver and lesions in CT abdomen images using cascaded fully convolutional neural networks (CFCNs) and dense 3D conditional random fields (CRFs). We train and cascade two FCNs for a combined segmentation of the liver and its lesions. In the first step, we train a FCN to segment the liver as ROI input for a second FCN. The second FCN solely segments lesions from the predicted liver ROIs of step 1. We refine the segmentations of the CFCN using a dense 3D CRF that accounts for both spatial coherence and appearance. CFCN models were trained in a 2-fold cross-validation on the abdominal CT dataset 3DIRCAD comprising 15 hepatic tumor volumes. Our results show that CFCN-based semantic liver and lesion segmentation achieves Dice scores over 94% for liver with computation times below 100s per volume. We experimentally demonstrate the robustness of the proposed method as a decision support system with a high accuracy and speed for usage in daily clinical routine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge