Seong Jae Hwang
Mono-Modalizing Extremely Heterogeneous Multi-Modal Medical Image Registration
Jun 18, 2025Abstract:In clinical practice, imaging modalities with functional characteristics, such as positron emission tomography (PET) and fractional anisotropy (FA), are often aligned with a structural reference (e.g., MRI, CT) for accurate interpretation or group analysis, necessitating multi-modal deformable image registration (DIR). However, due to the extreme heterogeneity of these modalities compared to standard structural scans, conventional unsupervised DIR methods struggle to learn reliable spatial mappings and often distort images. We find that the similarity metrics guiding these models fail to capture alignment between highly disparate modalities. To address this, we propose M2M-Reg (Multi-to-Mono Registration), a novel framework that trains multi-modal DIR models using only mono-modal similarity while preserving the established architectural paradigm for seamless integration into existing models. We also introduce GradCyCon, a regularizer that leverages M2M-Reg's cyclic training scheme to promote diffeomorphism. Furthermore, our framework naturally extends to a semi-supervised setting, integrating pre-aligned and unaligned pairs only, without requiring ground-truth transformations or segmentation masks. Experiments on the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset demonstrate that M2M-Reg achieves up to 2x higher DSC than prior methods for PET-MRI and FA-MRI registration, highlighting its effectiveness in handling highly heterogeneous multi-modal DIR. Our code is available at https://github.com/MICV-yonsei/M2M-Reg.
Backbone Augmented Training for Adaptations
Jun 04, 2025Abstract:Adaptations facilitate efficient training of large backbone models, including diffusion models for image generation and transformer-based language models. While various adaptation techniques enhance performance with minimal computational resources, limited adaptation data often leads to challenges in training. To address this, we focus on the enormous amount of backbone data used to pre-train the backbone models. We propose Backbone Augmented Training (BAT), a method that leverages backbone data to augment the adaptation dataset. First, we formulate and prove two mathematical key propositions: one establishes the validity of BAT, while the other identifies a condition under which BAT benefits adaptation. Furthermore, we introduce an advanced data selection scheme that satisfies these propositions and present ALBAT algorithm to implement this approach. ALBAT efficiently enhances adaptation training in both personalization and language generation tasks with scarce data.
PRETI: Patient-Aware Retinal Foundation Model via Metadata-Guided Representation Learning
May 18, 2025Abstract:Retinal foundation models have significantly advanced retinal image analysis by leveraging self-supervised learning to reduce dependence on labeled data while achieving strong generalization. Many recent approaches enhance retinal image understanding using report supervision, but obtaining clinical reports is often costly and challenging. In contrast, metadata (e.g., age, gender) is widely available and serves as a valuable resource for analyzing disease progression. To effectively incorporate patient-specific information, we propose PRETI, a retinal foundation model that integrates metadata-aware learning with robust self-supervised representation learning. We introduce Learnable Metadata Embedding (LME), which dynamically refines metadata representations. Additionally, we construct patient-level data pairs, associating images from the same individual to improve robustness against non-clinical variations. To further optimize retinal image representation, we propose Retina-Aware Adaptive Masking (RAAM), a strategy that selectively applies masking within the retinal region and dynamically adjusts the masking ratio during training. PRETI captures both global structures and fine-grained pathological details, resulting in superior diagnostic performance. Extensive experiments demonstrate that PRETI achieves state-of-the-art results across diverse diseases and biomarker predictions using in-house and public data, indicating the importance of metadata-guided foundation models in retinal disease analysis. Our code and pretrained model are available at https://github.com/MICV-yonsei/PRETI
Spatial Transport Optimization by Repositioning Attention Map for Training-Free Text-to-Image Synthesis
Mar 28, 2025



Abstract:Diffusion-based text-to-image (T2I) models have recently excelled in high-quality image generation, particularly in a training-free manner, enabling cost-effective adaptability and generalization across diverse tasks. However, while the existing methods have been continuously focusing on several challenges, such as "missing objects" and "mismatched attributes," another critical issue of "mislocated objects" remains where generated spatial positions fail to align with text prompts. Surprisingly, ensuring such seemingly basic functionality remains challenging in popular T2I models due to the inherent difficulty of imposing explicit spatial guidance via text forms. To address this, we propose STORM (Spatial Transport Optimization by Repositioning Attention Map), a novel training-free approach for spatially coherent T2I synthesis. STORM employs Spatial Transport Optimization (STO), rooted in optimal transport theory, to dynamically adjust object attention maps for precise spatial adherence, supported by a Spatial Transport (ST) Cost function that enhances spatial understanding. Our analysis shows that integrating spatial awareness is most effective in the early denoising stages, while later phases refine details. Extensive experiments demonstrate that STORM surpasses existing methods, effectively mitigating mislocated objects while improving missing and mismatched attributes, setting a new benchmark for spatial alignment in T2I synthesis.
Rethinking Glaucoma Calibration: Voting-Based Binocular and Metadata Integration
Mar 24, 2025Abstract:Glaucoma is an incurable ophthalmic disease that damages the optic nerve, leads to vision loss, and ranks among the leading causes of blindness worldwide. Diagnosing glaucoma typically involves fundus photography, optical coherence tomography (OCT), and visual field testing. However, the high cost of OCT often leads to reliance on fundus photography and visual field testing, both of which exhibit inherent inter-observer variability. This stems from glaucoma being a multifaceted disease that influenced by various factors. As a result, glaucoma diagnosis is highly subjective, emphasizing the necessity of calibration, which aligns predicted probabilities with actual disease likelihood. Proper calibration is essential to prevent overdiagnosis or misdiagnosis, which are critical concerns for high-risk diseases. Although AI has significantly improved diagnostic accuracy, overconfidence in models have worsen calibration performance. Recent study has begun focusing on calibration for glaucoma. Nevertheless, previous study has not fully considered glaucoma's systemic nature and the high subjectivity in its diagnostic process. To overcome these limitations, we propose V-ViT (Voting-based ViT), a novel framework that enhances calibration by incorporating disease-specific characteristics. V-ViT integrates binocular data and metadata, reflecting the multi-faceted nature of glaucoma diagnosis. Additionally, we introduce a MC dropout-based Voting System to address high subjectivity. Our approach achieves state-of-the-art performance across all metrics, including accuracy, demonstrating that our proposed methods are effective in addressing calibration issues. We validate our method using a custom dataset including binocular data.
Fourier Decomposition for Explicit Representation of 3D Point Cloud Attributes
Mar 13, 2025Abstract:While 3D point clouds are widely utilized across various vision applications, their irregular and sparse nature make them challenging to handle. In response, numerous encoding approaches have been proposed to capture the rich semantic information of point clouds. Yet, a critical limitation persists: a lack of consideration for colored point clouds which are more capable 3D representations as they contain diverse attributes: color and geometry. While existing methods handle these attributes separately on a per-point basis, this leads to a limited receptive field and restricted ability to capture relationships across multiple points. To address this, we pioneer a point cloud encoding methodology that leverages 3D Fourier decomposition to disentangle color and geometric features while extending the receptive field through spectral-domain operations. Our analysis confirms that this encoding approach effectively separates feature components, where the amplitude uniquely captures color attributes and the phase encodes geometric structure, thereby enabling independent learning and utilization of both attributes. Furthermore, the spectral-domain properties of these components naturally aggregate local features while considering multiple points' information. We validate our point cloud encoding approach on point cloud classification and style transfer tasks, achieving state-of-the-art results on the DensePoint dataset with improvements via a proposed amplitude-based data augmentation strategy.
Pathology-Aware Adaptive Watermarking for Text-Driven Medical Image Synthesis
Mar 11, 2025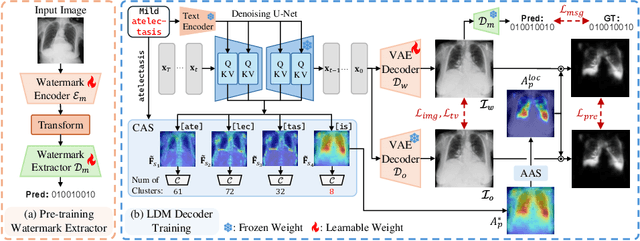
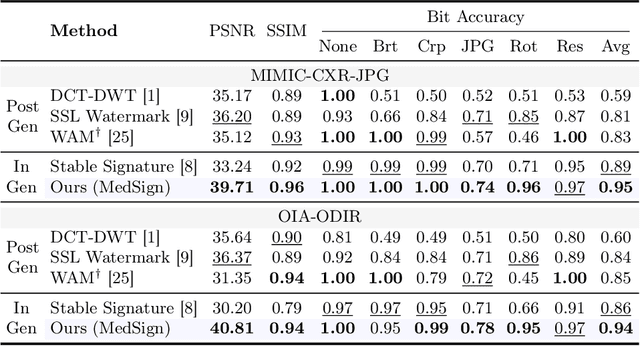
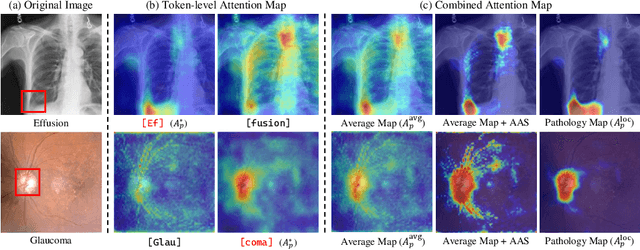
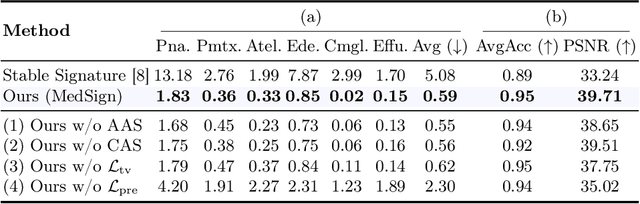
Abstract:As recent text-conditioned diffusion models have enabled the generation of high-quality images, concerns over their potential misuse have also grown. This issue is critical in the medical domain, where text-conditioned generated medical images could enable insurance fraud or falsified records, highlighting the urgent need for reliable safeguards against unethical use. While watermarking techniques have emerged as a promising solution in general image domains, their direct application to medical imaging presents significant challenges. A key challenge is preserving fine-grained disease manifestations, as even minor distortions from a watermark may lead to clinical misinterpretation, which compromises diagnostic integrity. To overcome this gap, we present MedSign, a deep learning-based watermarking framework specifically designed for text-to-medical image synthesis, which preserves pathologically significant regions by adaptively adjusting watermark strength. Specifically, we generate a pathology localization map using cross-attention between medical text tokens and the diffusion denoising network, aggregating token-wise attention across layers, heads, and time steps. Leveraging this map, we optimize the LDM decoder to incorporate watermarking during image synthesis, ensuring cohesive integration while minimizing interference in diagnostically critical regions. Experimental results show that our MedSign preserves diagnostic integrity while ensuring watermark robustness, achieving state-of-the-art performance in image quality and detection accuracy on MIMIC-CXR and OIA-ODIR datasets.
Your Large Vision-Language Model Only Needs A Few Attention Heads For Visual Grounding
Mar 08, 2025Abstract:Visual grounding seeks to localize the image region corresponding to a free-form text description. Recently, the strong multimodal capabilities of Large Vision-Language Models (LVLMs) have driven substantial improvements in visual grounding, though they inevitably require fine-tuning and additional model components to explicitly generate bounding boxes or segmentation masks. However, we discover that a few attention heads in frozen LVLMs demonstrate strong visual grounding capabilities. We refer to these heads, which consistently capture object locations related to text semantics, as localization heads. Using localization heads, we introduce a straightforward and effective training-free visual grounding framework that utilizes text-to-image attention maps from localization heads to identify the target objects. Surprisingly, only three out of thousands of attention heads are sufficient to achieve competitive localization performance compared to existing LVLM-based visual grounding methods that require fine-tuning. Our findings suggest that LVLMs can innately ground objects based on a deep comprehension of the text-image relationship, as they implicitly focus on relevant image regions to generate informative text outputs. All the source codes will be made available to the public.
See What You Are Told: Visual Attention Sink in Large Multimodal Models
Mar 05, 2025Abstract:Large multimodal models (LMMs) "see" images by leveraging the attention mechanism between text and visual tokens in the transformer decoder. Ideally, these models should focus on key visual information relevant to the text token. However, recent findings indicate that LMMs have an extraordinary tendency to consistently allocate high attention weights to specific visual tokens, even when these tokens are irrelevant to the corresponding text. In this study, we investigate the property behind the appearance of these irrelevant visual tokens and examine their characteristics. Our findings show that this behavior arises due to the massive activation of certain hidden state dimensions, which resembles the attention sink found in language models. Hence, we refer to this phenomenon as the visual attention sink. In particular, our analysis reveals that removing the irrelevant visual sink tokens does not impact model performance, despite receiving high attention weights. Consequently, we recycle the attention to these tokens as surplus resources, redistributing the attention budget to enhance focus on the image. To achieve this, we introduce Visual Attention Redistribution (VAR), a method that redistributes attention in image-centric heads, which we identify as innately focusing on visual information. VAR can be seamlessly applied across different LMMs to improve performance on a wide range of tasks, including general vision-language tasks, visual hallucination tasks, and vision-centric tasks, all without the need for additional training, models, or inference steps. Experimental results demonstrate that VAR enables LMMs to process visual information more effectively by adjusting their internal attention mechanisms, offering a new direction to enhancing the multimodal capabilities of LMMs.
Distilling Spectral Graph for Object-Context Aware Open-Vocabulary Semantic Segmentation
Nov 26, 2024Abstract:Open-Vocabulary Semantic Segmentation (OVSS) has advanced with recent vision-language models (VLMs), enabling segmentation beyond predefined categories through various learning schemes. Notably, training-free methods offer scalable, easily deployable solutions for handling unseen data, a key goal of OVSS. Yet, a critical issue persists: lack of object-level context consideration when segmenting complex objects in the challenging environment of OVSS based on arbitrary query prompts. This oversight limits models' ability to group semantically consistent elements within object and map them precisely to user-defined arbitrary classes. In this work, we introduce a novel approach that overcomes this limitation by incorporating object-level contextual knowledge within images. Specifically, our model enhances intra-object consistency by distilling spectral-driven features from vision foundation models into the attention mechanism of the visual encoder, enabling semantically coherent components to form a single object mask. Additionally, we refine the text embeddings with zero-shot object presence likelihood to ensure accurate alignment with the specific objects represented in the images. By leveraging object-level contextual knowledge, our proposed approach achieves state-of-the-art performance with strong generalizability across diverse datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge