Randal Burns
Johns Hopkins University
Auditing Significance, Metric Choice, and Demographic Fairness in Medical AI Challenges
Dec 22, 2025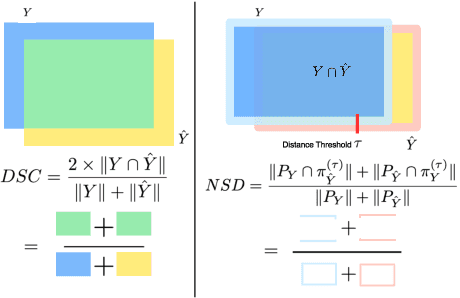
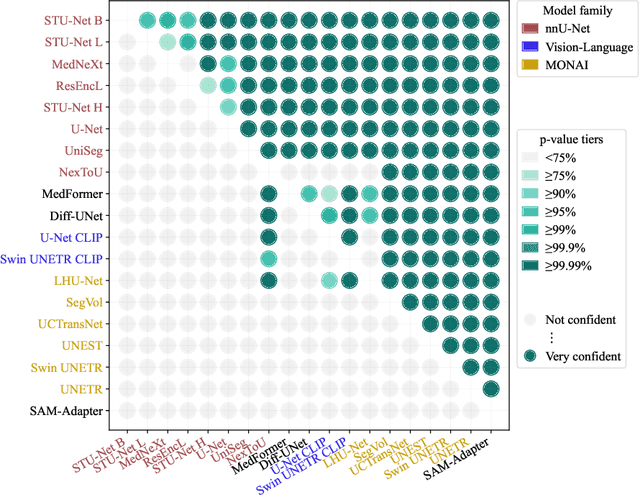
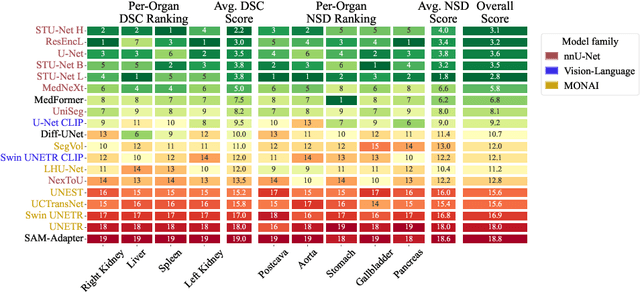
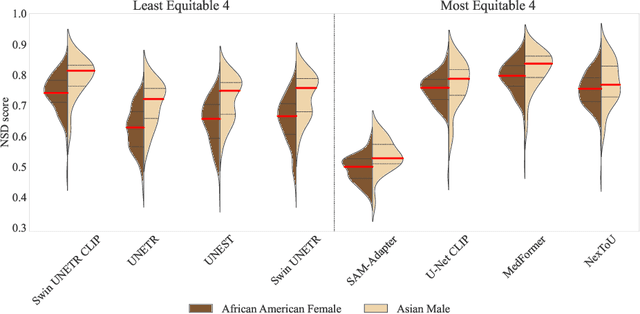
Abstract:Open challenges have become the de facto standard for comparative ranking of medical AI methods. Despite their importance, medical AI leaderboards exhibit three persistent limitations: (1) score gaps are rarely tested for statistical significance, so rank stability is unknown; (2) single averaged metrics are applied to every organ, hiding clinically important boundary errors; (3) performance across intersecting demographics is seldom reported, masking fairness and equity gaps. We introduce RankInsight, an open-source toolkit that seeks to address these limitations. RankInsight (1) computes pair-wise significance maps that show the nnU-Net family outperforms Vision-Language and MONAI submissions with high statistical certainty; (2) recomputes leaderboards with organ-appropriate metrics, reversing the order of the top four models when Dice is replaced by NSD for tubular structures; and (3) audits intersectional fairness, revealing that more than half of the MONAI-based entries have the largest gender-race discrepancy on our proprietary Johns Hopkins Hospital dataset. The RankInsight toolkit is publicly released and can be directly applied to past, ongoing, and future challenges. It enables organizers and participants to publish rankings that are statistically sound, clinically meaningful, and demographically fair.
On Harnessing Idle Compute at the Edge for Foundation Model Training
Dec 13, 2025Abstract:The ecosystem behind foundation model development today is highly centralized and limited to large-scale cloud data center operators: training foundation models is costly, needing immense compute resources. Decentralized foundation model training across edge devices, leveraging their spare compute, promises a democratized alternative. However, existing edge-training approaches fall short: they struggle to match cloud-based training performance, exhibit limited scalability with model size, exceed device memory capacity, and have prohibitive communication overhead. They also fail to satisfactorily handle device heterogeneity and dynamism. We introduce a new paradigm, Cleave, which finely partitions training operations through a novel selective hybrid tensor parallelism method. Together with a parameter server centric training framework, Cleave copes with device memory limits and avoids communication bottlenecks, thereby enabling efficient training of large models on par with the cloud. Further, with a cost optimization model to guide device selection and training workload distribution, Cleave effectively accounts for device heterogeneity and churn. Our evaluations show that Cleave matches cloud-based GPU training by scaling efficiently to larger models and thousands of devices, supporting up to 8x more devices than baseline edge-training approaches. It outperforms state-of-the-art edge training methods by up to a factor of 10 in per-batch training time and efficiently handles device failures, achieving at least 100x faster recovery than prior methods.
Towards Decentralized and Sustainable Foundation Model Training with the Edge
Jul 02, 2025Abstract:Foundation models are at the forefront of AI research, appealing for their ability to learn from vast datasets and cater to diverse tasks. Yet, their significant computational demands raise issues of environmental impact and the risk of centralized control in their development. We put forward a vision towards decentralized and sustainable foundation model training that leverages the collective compute of sparingly used connected edge AI devices. We present the rationale behind our vision, particularly in support of its sustainability benefit. We further outline a set of challenges that need to be addressed to turn this vision into reality.
Masked Matrix Multiplication for Emergent Sparsity
Feb 21, 2024



Abstract:Artificial intelligence workloads, especially transformer models, exhibit emergent sparsity in which computations perform selective sparse access to dense data. The workloads are inefficient on hardware designed for dense computations and do not map well onto sparse data representations. We build a vectorized and parallel matrix-multiplication system A X B = C that eliminates unnecessary computations and avoids branches based on a runtime evaluation of sparsity. We use a combination of dynamic code lookup to adapt to the specific sparsity encoded in the B matrix and preprocessing of sparsity maps of the A and B matrices to compute conditional branches once for the whole computation. For a wide range of sparsity, from 60% to 95% zeros, our implementation performs fewer instructions and increases performance when compared with Intel MKL's dense or sparse matrix multiply routines. Benefits can be as large as 2 times speedup and 4 times fewer instructions.
Edge-Parallel Graph Encoder Embedding
Feb 06, 2024



Abstract:New algorithms for embedding graphs have reduced the asymptotic complexity of finding low-dimensional representations. One-Hot Graph Encoder Embedding (GEE) uses a single, linear pass over edges and produces an embedding that converges asymptotically to the spectral embedding. The scaling and performance benefits of this approach have been limited by a serial implementation in an interpreted language. We refactor GEE into a parallel program in the Ligra graph engine that maps functions over the edges of the graph and uses lock-free atomic instrutions to prevent data races. On a graph with 1.8B edges, this results in a 500 times speedup over the original implementation and a 17 times speedup over a just-in-time compiled version.
Understanding Patterns of Deep Learning ModelEvolution in Network Architecture Search
Sep 22, 2023Abstract:Network Architecture Search and specifically Regularized Evolution is a common way to refine the structure of a deep learning model.However, little is known about how models empirically evolve over time which has design implications for designing caching policies, refining the search algorithm for particular applications, and other important use cases.In this work, we algorithmically analyze and quantitatively characterize the patterns of model evolution for a set of models from the Candle project and the Nasbench-201 search space.We show how the evolution of the model structure is influenced by the regularized evolution algorithm. We describe how evolutionary patterns appear in distributed settings and opportunities for caching and improved scheduling. Lastly, we describe the conditions that affect when particular model architectures rise and fall in popularity based on their frequency of acting as a donor in a sliding window.
Prospective Learning: Back to the Future
Jan 19, 2022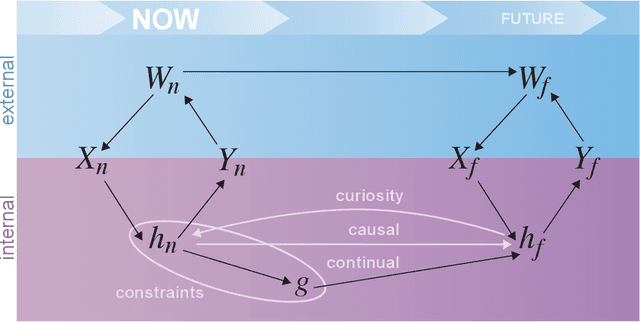
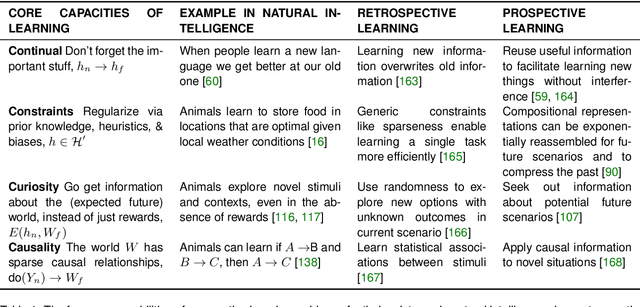
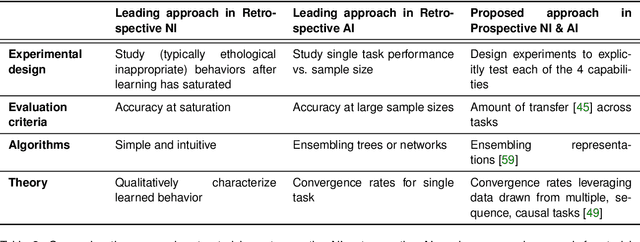
Abstract:Research on both natural intelligence (NI) and artificial intelligence (AI) generally assumes that the future resembles the past: intelligent agents or systems (what we call 'intelligence') observe and act on the world, then use this experience to act on future experiences of the same kind. We call this 'retrospective learning'. For example, an intelligence may see a set of pictures of objects, along with their names, and learn to name them. A retrospective learning intelligence would merely be able to name more pictures of the same objects. We argue that this is not what true intelligence is about. In many real world problems, both NIs and AIs will have to learn for an uncertain future. Both must update their internal models to be useful for future tasks, such as naming fundamentally new objects and using these objects effectively in a new context or to achieve previously unencountered goals. This ability to learn for the future we call 'prospective learning'. We articulate four relevant factors that jointly define prospective learning. Continual learning enables intelligences to remember those aspects of the past which it believes will be most useful in the future. Prospective constraints (including biases and priors) facilitate the intelligence finding general solutions that will be applicable to future problems. Curiosity motivates taking actions that inform future decision making, including in previously unmet situations. Causal estimation enables learning the structure of relations that guide choosing actions for specific outcomes, even when the specific action-outcome contingencies have never been observed before. We argue that a paradigm shift from retrospective to prospective learning will enable the communities that study intelligence to unite and overcome existing bottlenecks to more effectively explain, augment, and engineer intelligences.
PACSET (Packed Serialized Trees): Reducing Inference Latency for Tree Ensemble Deployment
Nov 10, 2020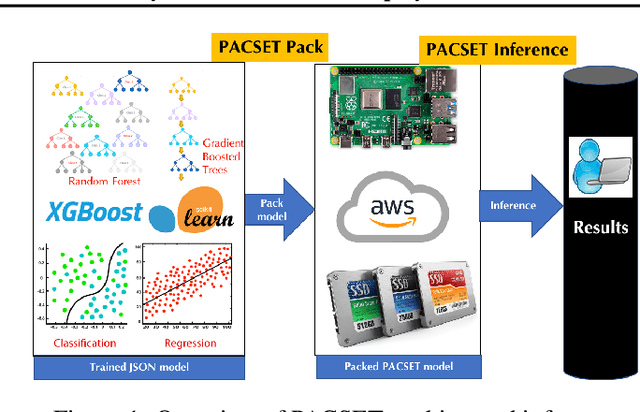


Abstract:We present methods to serialize and deserialize tree ensembles that optimize inference latency when models are not already loaded into memory. This arises whenever models are larger than memory, but also systematically when models are deployed on low-resource devices, such as in the Internet of Things, or run as Web micro-services where resources are allocated on demand. Our packed serialized trees (PACSET) encode reference locality in the layout of a tree ensemble using principles from external memory algorithms. The layout interleaves correlated nodes across multiple trees, uses leaf cardinality to collocate the nodes on the most popular paths and is optimized for the I/O blocksize. The result is that each I/O yields a higher fraction of useful data, leading to a 2-6 times reduction in classification latency for interactive workloads.
Geodesic Learning via Unsupervised Decision Forests
Jul 05, 2019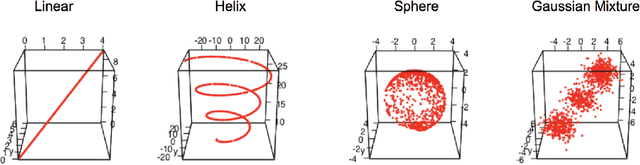
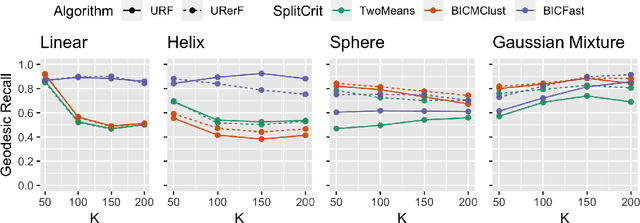
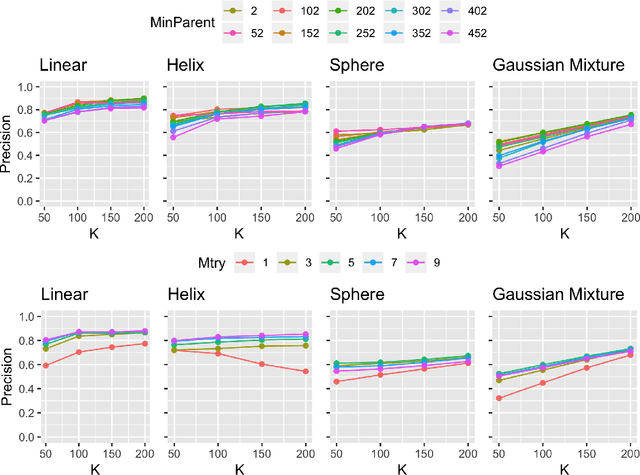
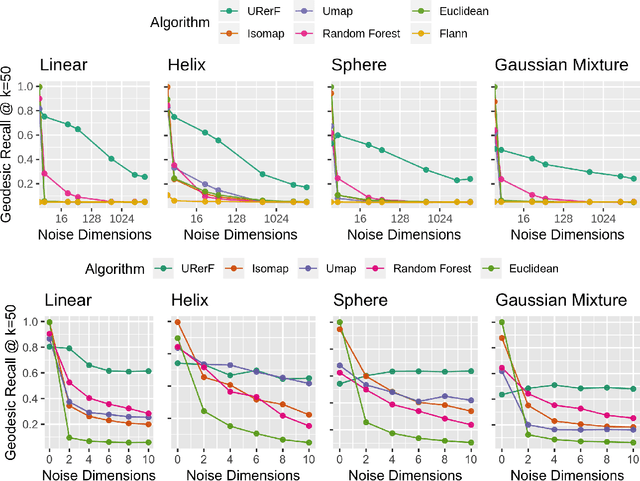
Abstract:Geodesic distance is the shortest path between two points in a Riemannian manifold. Manifold learning algorithms, such as Isomap, seek to learn a manifold that preserves geodesic distances. However, such methods operate on the ambient dimensionality, and are therefore fragile to noise dimensions. We developed an unsupervised random forest method (URerF) to approximately learn geodesic distances in linear and nonlinear manifolds with noise. URerF operates on low-dimensional sparse linear combinations of features, rather than the full observed dimensionality. To choose the optimal split in a computationally efficient fashion, we developed a fast Bayesian Information Criterion statistic for Gaussian mixture models. We introduce geodesic precision-recall curves which quantify performance relative to the true latent manifold. Empirical results on simulated and real data demonstrate that URerF is robust to high-dimensional noise, where as other methods, such as Isomap, UMAP, and FLANN, quickly deteriorate in such settings. In particular, URerF is able to estimate geodesic distances on a real connectome dataset better than other approaches.
Random Projection Forests
Oct 10, 2018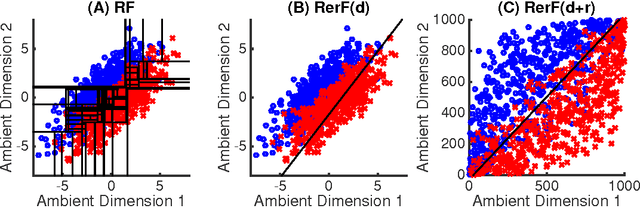
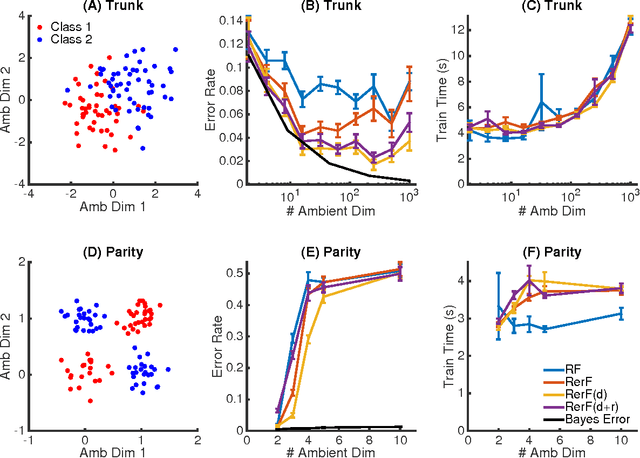
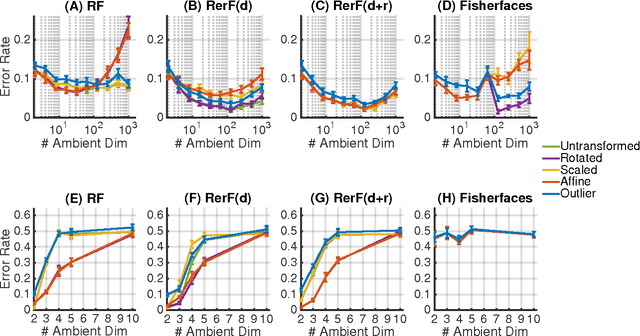
Abstract:Ensemble methods---particularly those based on decision trees---have recently demonstrated superior performance in a variety of machine learning settings. We introduce a generalization of many existing decision tree methods called "Random Projection Forests" (RPF), which is any decision forest that uses (possibly data dependent and random) linear projections. Using this framework, we introduce a special case, called "Lumberjack", using very sparse random projections, that is, linear combinations of a small subset of features. Lumberjack obtains statistically significantly improved accuracy over Random Forests, Gradient Boosted Trees, and other approaches on a standard benchmark suites for classification with varying dimension, sample size, and number of classes. To illustrate how, why, and when Lumberjack outperforms other methods, we conduct extensive simulated experiments, in vectors, images, and nonlinear manifolds. Lumberjack typically yields improved performance over existing decision trees ensembles, while mitigating computational efficiency and scalability, and maintaining interpretability. Lumberjack can easily be incorporated into other ensemble methods such as boosting to obtain potentially similar gains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge