Paul M. Thompson
for the Alzheimer's Disease Neuroimaging Initiative
Improved Prediction of Beta-Amyloid and Tau Burden Using Hippocampal Surface Multivariate Morphometry Statistics and Sparse Coding
Oct 28, 2022Abstract:Background: Beta-amyloid (A$\beta$) plaques and tau protein tangles in the brain are the defining 'A' and 'T' hallmarks of Alzheimer's disease (AD), and together with structural atrophy detectable on brain magnetic resonance imaging (MRI) scans as one of the neurodegenerative ('N') biomarkers comprise the ''ATN framework'' of AD. Current methods to detect A$\beta$/tau pathology include cerebrospinal fluid (CSF; invasive), positron emission tomography (PET; costly and not widely available), and blood-based biomarkers (BBBM; promising but mainly still in development). Objective: To develop a non-invasive and widely available structural MRI-based framework to quantitatively predict the amyloid and tau measurements. Methods: With MRI-based hippocampal multivariate morphometry statistics (MMS) features, we apply our Patch Analysis-based Surface Correntropy-induced Sparse coding and max-pooling (PASCS-MP) method combined with the ridge regression model to individual amyloid/tau measure prediction. Results: We evaluate our framework on amyloid PET/MRI and tau PET/MRI datasets from the Alzheimer's Disease Neuroimaging Initiative (ADNI). Each subject has one pair consisting of a PET image and MRI scan, collected at about the same time. Experimental results suggest that amyloid/tau measurements predicted with our PASCP-MP representations are closer to the real values than the measures derived from other approaches, such as hippocampal surface area, volume, and shape morphometry features based on spherical harmonics (SPHARM). Conclusion: The MMS-based PASCP-MP is an efficient tool that can bridge hippocampal atrophy with amyloid and tau pathology and thus help assess disease burden, progression, and treatment effects.
Secure Federated Learning for Neuroimaging
May 11, 2022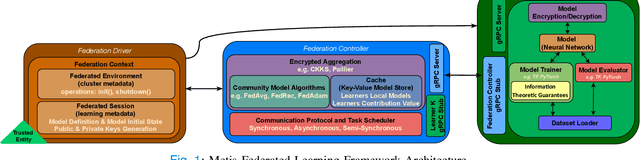
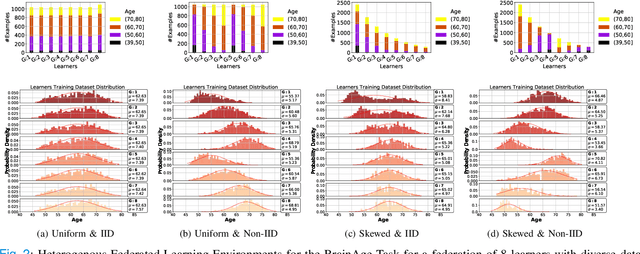
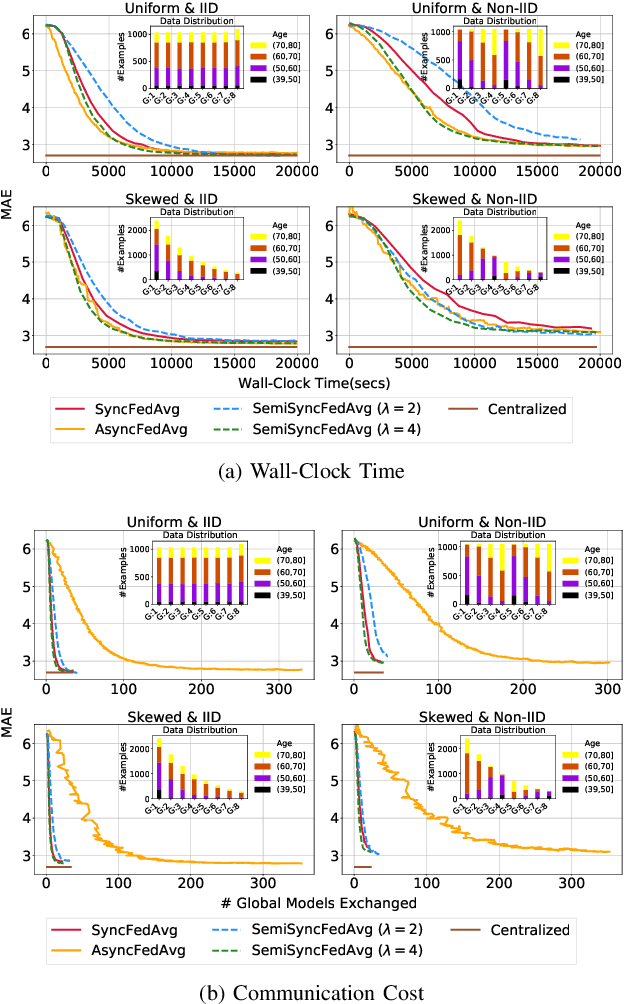
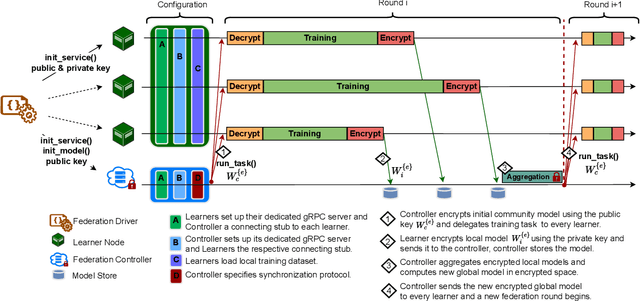
Abstract:The amount of biomedical data continues to grow rapidly. However, the ability to collect data from multiple sites for joint analysis remains challenging due to security, privacy, and regulatory concerns. We present a Secure Federated Learning architecture, MetisFL, which enables distributed training of neural networks over multiple data sources without sharing data. Each site trains the neural network over its private data for some time, then shares the neural network parameters (i.e., weights, gradients) with a Federation Controller, which in turn aggregates the local models, sends the resulting community model back to each site, and the process repeats. Our architecture provides strong security and privacy. First, sample data never leaves a site. Second, neural parameters are encrypted before transmission and the community model is computed under fully-homomorphic encryption. Finally, we use information-theoretic methods to limit information leakage from the neural model to prevent a curious site from performing membership attacks. We demonstrate this architecture in neuroimaging. Specifically, we investigate training neural models to classify Alzheimer's disease, and estimate Brain Age, from magnetic resonance imaging datasets distributed across multiple sites, including heterogeneous environments where sites have different amounts of data, statistical distributions, and computational capabilities.
Predicting Tau Accumulation in Cerebral Cortex with Multivariate MRI Morphometry Measurements, Sparse Coding, and Correntropy
Oct 20, 2021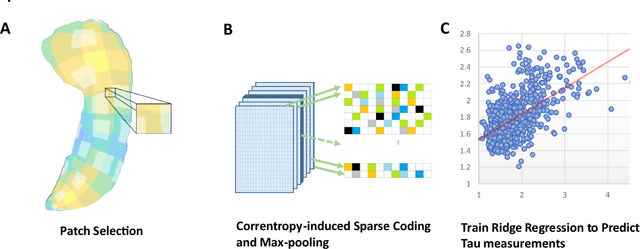
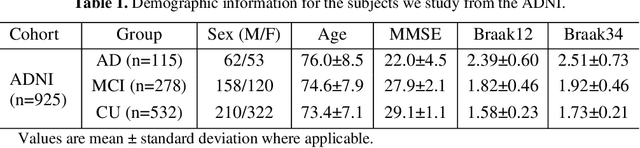
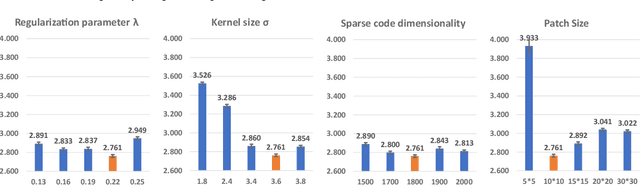
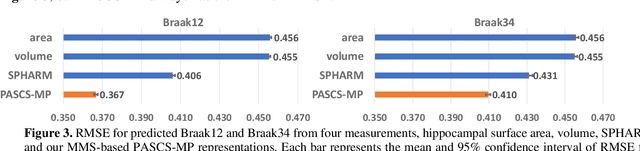
Abstract:Biomarker-assisted diagnosis and intervention in Alzheimer's disease (AD) may be the key to prevention breakthroughs. One of the hallmarks of AD is the accumulation of tau plaques in the human brain. However, current methods to detect tau pathology are either invasive (lumbar puncture) or quite costly and not widely available (Tau PET). In our previous work, structural MRI-based hippocampal multivariate morphometry statistics (MMS) showed superior performance as an effective neurodegenerative biomarker for preclinical AD and Patch Analysis-based Surface Correntropy-induced Sparse coding and max-pooling (PASCS-MP) has excellent ability to generate low-dimensional representations with strong statistical power for brain amyloid prediction. In this work, we apply this framework together with ridge regression models to predict Tau deposition in Braak12 and Braak34 brain regions separately. We evaluate our framework on 925 subjects from the Alzheimer's Disease Neuroimaging Initiative (ADNI). Each subject has one pair consisting of a PET image and MRI scan which were collected at about the same times. Experimental results suggest that the representations from our MMS and PASCS-MP have stronger predictive power and their predicted Braak12 and Braak34 are closer to the real values compared to the measures derived from other approaches such as hippocampal surface area and volume, and shape morphometry features based on spherical harmonics (SPHARM).
Secure Neuroimaging Analysis using Federated Learning with Homomorphic Encryption
Aug 07, 2021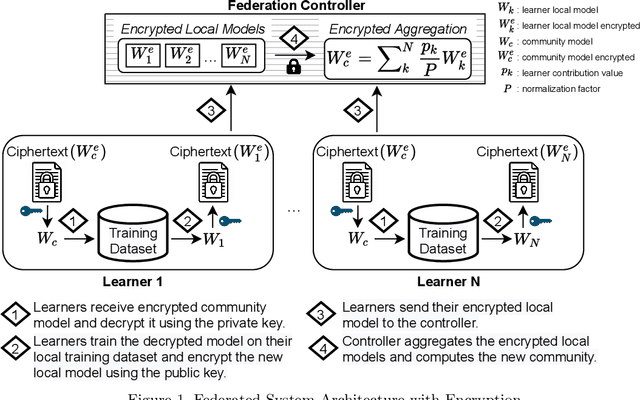
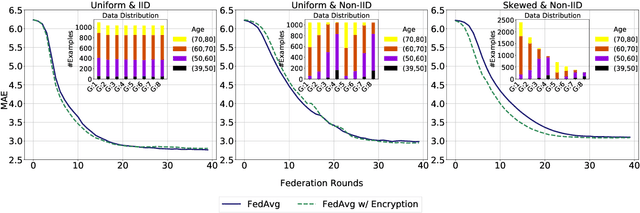
Abstract:Federated learning (FL) enables distributed computation of machine learning models over various disparate, remote data sources, without requiring to transfer any individual data to a centralized location. This results in an improved generalizability of models and efficient scaling of computation as more sources and larger datasets are added to the federation. Nevertheless, recent membership attacks show that private or sensitive personal data can sometimes be leaked or inferred when model parameters or summary statistics are shared with a central site, requiring improved security solutions. In this work, we propose a framework for secure FL using fully-homomorphic encryption (FHE). Specifically, we use the CKKS construction, an approximate, floating point compatible scheme that benefits from ciphertext packing and rescaling. In our evaluation on large-scale brain MRI datasets, we use our proposed secure FL framework to train a deep learning model to predict a person's age from distributed MRI scans, a common benchmarking task, and demonstrate that there is no degradation in the learning performance between the encrypted and non-encrypted federated models.
Membership Inference Attacks on Deep Regression Models for Neuroimaging
Jun 03, 2021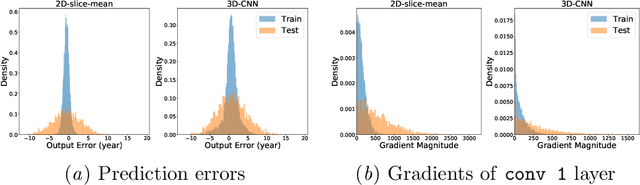


Abstract:Ensuring the privacy of research participants is vital, even more so in healthcare environments. Deep learning approaches to neuroimaging require large datasets, and this often necessitates sharing data between multiple sites, which is antithetical to the privacy objectives. Federated learning is a commonly proposed solution to this problem. It circumvents the need for data sharing by sharing parameters during the training process. However, we demonstrate that allowing access to parameters may leak private information even if data is never directly shared. In particular, we show that it is possible to infer if a sample was used to train the model given only access to the model prediction (black-box) or access to the model itself (white-box) and some leaked samples from the training data distribution. Such attacks are commonly referred to as Membership Inference attacks. We show realistic Membership Inference attacks on deep learning models trained for 3D neuroimaging tasks in a centralized as well as decentralized setup. We demonstrate feasible attacks on brain age prediction models (deep learning models that predict a person's age from their brain MRI scan). We correctly identified whether an MRI scan was used in model training with a 60% to over 80% success rate depending on model complexity and security assumptions.
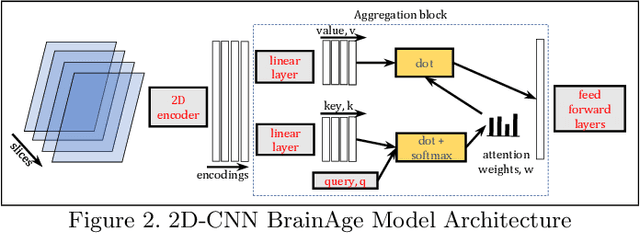
Improved Brain Age Estimation with Slice-based Set Networks
Feb 09, 2021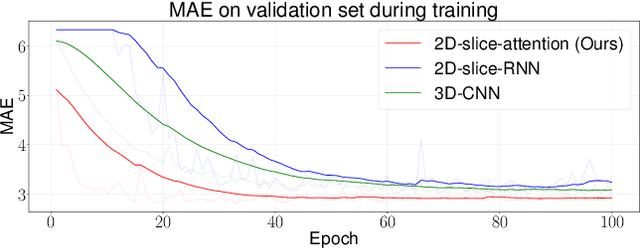
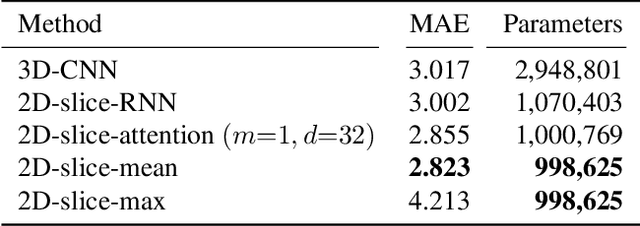
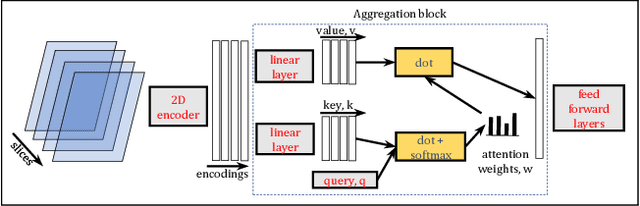
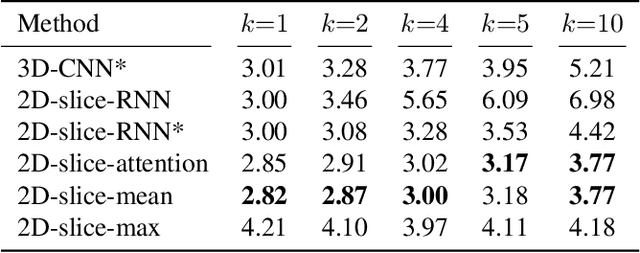
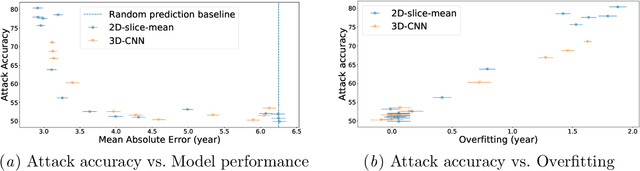
Abstract:Deep Learning for neuroimaging data is a promising but challenging direction. The high dimensionality of 3D MRI scans makes this endeavor compute and data-intensive. Most conventional 3D neuroimaging methods use 3D-CNN-based architectures with a large number of parameters and require more time and data to train. Recently, 2D-slice-based models have received increasing attention as they have fewer parameters and may require fewer samples to achieve comparable performance. In this paper, we propose a new architecture for BrainAGE prediction. The proposed architecture works by encoding each 2D slice in an MRI with a deep 2D-CNN model. Next, it combines the information from these 2D-slice encodings using set networks or permutation invariant layers. Experiments on the BrainAGE prediction problem, using the UK Biobank dataset, showed that the model with the permutation invariant layers trains faster and provides better predictions compared to other state-of-the-art approaches.
Overview of Scanner Invariant Representations
May 29, 2020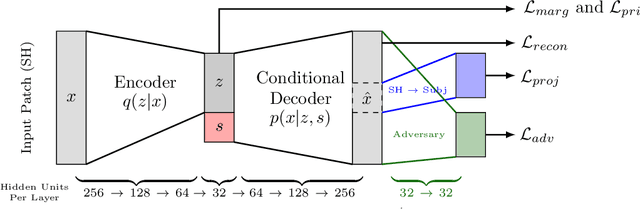
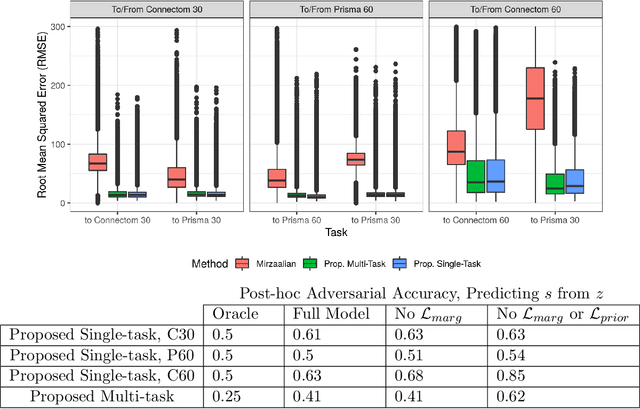
Abstract:Pooled imaging data from multiple sources is subject to bias from each source. Studies that do not correct for these scanner/site biases at best lose statistical power, and at worst leave spurious correlations in their data. Estimation of the bias effects is non-trivial due to the paucity of data with correspondence across sites, so called "traveling phantom" data, which is expensive to collect. Nevertheless, numerous solutions leveraging direct correspondence have been proposed. In contrast to this, Moyer et al. (2019) proposes an unsupervised solution using invariant representations, one which does not require correspondence and thus does not require paired images. By leveraging the data processing inequality, an invariant representation can then be used to create an image reconstruction that is uninformative of its original source, yet still faithful to the underlying structure. In the present abstract we provide an overview of this method.
Scanner Invariant Representations for Diffusion MRI Harmonization
Apr 10, 2019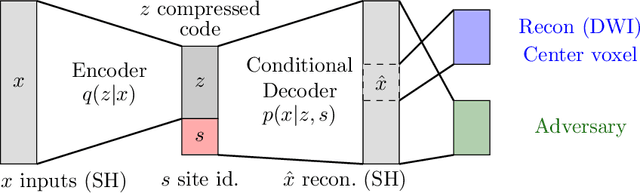
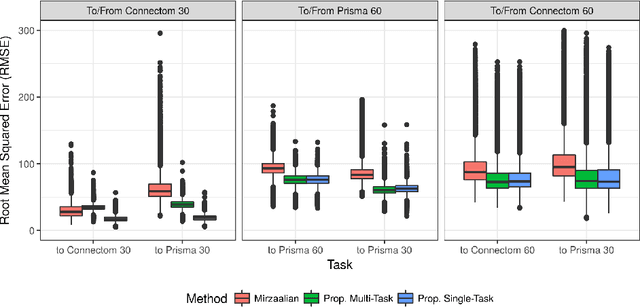
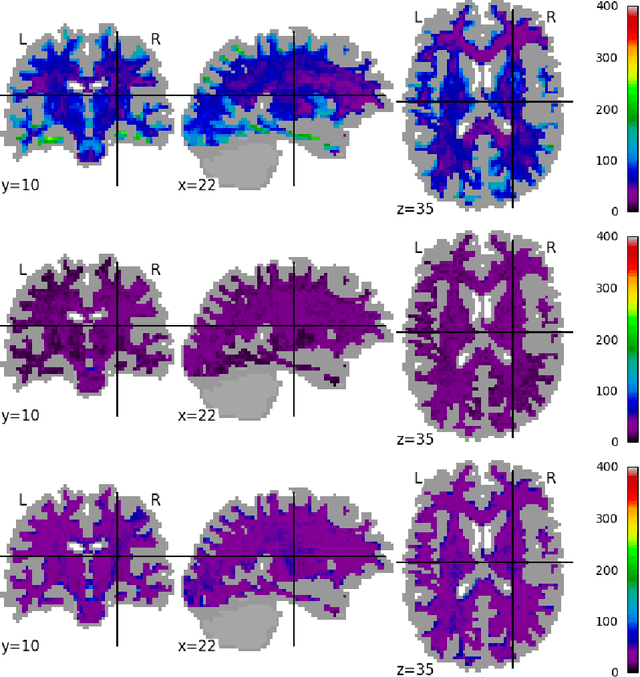
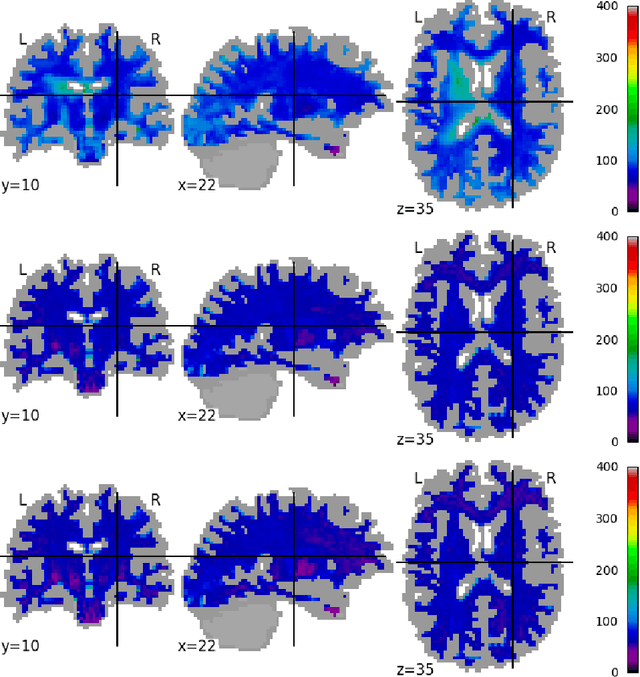
Abstract:Pooled imaging data from multiple sources is subject to variation between the sources. Correcting for these biases has become incredibly important as the size of imaging studies increases and the multi-site case becomes more common. We propose learning an intermediate representation invariant to site/protocol variables, a technique adapted from information theory-based algorithmic fairness; by leveraging the data processing inequality, such a representation can then be used to create an image reconstruction that is uninformative of its original source, yet still faithful to the underlying structure. To implement this, we use a machine learning method based on variational auto-encoders (VAE) to construct scanner invariant encodings of the imaging data. To evaluate our method, we use training data from the 2018 CDMRI Challenge Harmonization dataset. Our proposed method shows improvements on independent test data relative to a recently published baseline method.
Measures of Tractography Convergence
Jun 12, 2018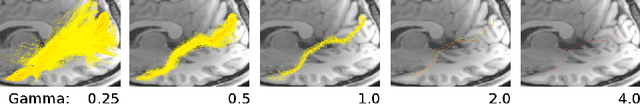
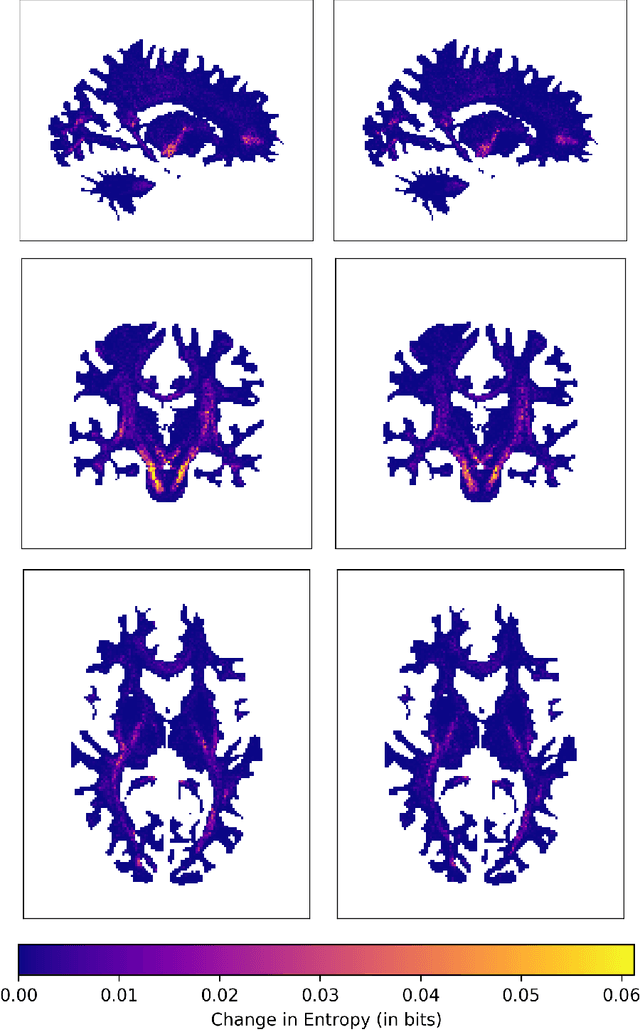
Abstract:In the present work, we use information theory to understand the empirical convergence rate of tractography, a widely-used approach to reconstruct anatomical fiber pathways in the living brain. Based on diffusion MRI data, tractography is the starting point for many methods to study brain connectivity. Of the available methods to perform tractography, most reconstruct a finite set of streamlines, or 3D curves, representing probable connections between anatomical regions, yet relatively little is known about how the sampling of this set of streamlines affects downstream results, and how exhaustive the sampling should be. Here we provide a method to measure the information theoretic surprise (self-cross entropy) for tract sampling schema. We then empirically assess four streamline methods. We demonstrate that the relative information gain is very low after a moderate number of streamlines have been generated for each tested method. The results give rise to several guidelines for optimal sampling in brain connectivity analyses.
Simultaneous Matrix Diagonalization for Structural Brain Networks Classification
Oct 14, 2017
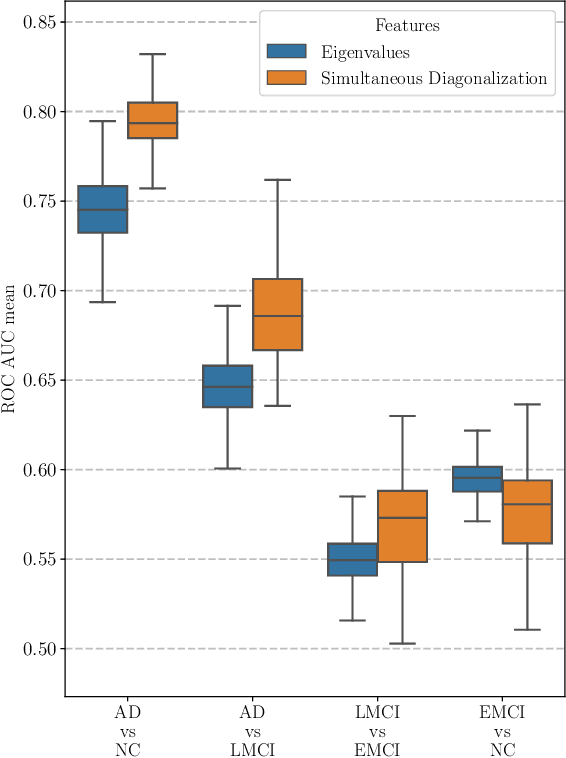

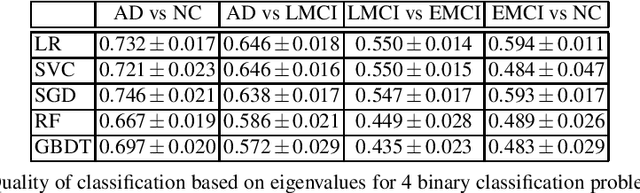
Abstract:This paper considers the problem of brain disease classification based on connectome data. A connectome is a network representation of a human brain. The typical connectome classification problem is very challenging because of the small sample size and high dimensionality of the data. We propose to use simultaneous approximate diagonalization of adjacency matrices in order to compute their eigenstructures in more stable way. The obtained approximate eigenvalues are further used as features for classification. The proposed approach is demonstrated to be efficient for detection of Alzheimer's disease, outperforming simple baselines and competing with state-of-the-art approaches to brain disease classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge