Pradeep K. Lam
Membership Inference Attacks on Deep Regression Models for Neuroimaging
Jun 03, 2021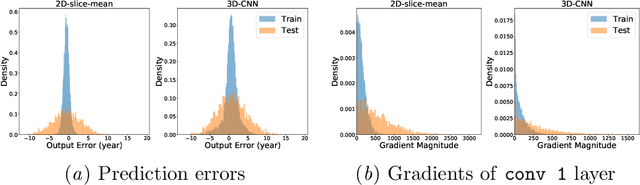


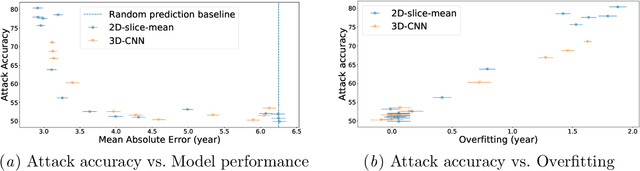
Abstract:Ensuring the privacy of research participants is vital, even more so in healthcare environments. Deep learning approaches to neuroimaging require large datasets, and this often necessitates sharing data between multiple sites, which is antithetical to the privacy objectives. Federated learning is a commonly proposed solution to this problem. It circumvents the need for data sharing by sharing parameters during the training process. However, we demonstrate that allowing access to parameters may leak private information even if data is never directly shared. In particular, we show that it is possible to infer if a sample was used to train the model given only access to the model prediction (black-box) or access to the model itself (white-box) and some leaked samples from the training data distribution. Such attacks are commonly referred to as Membership Inference attacks. We show realistic Membership Inference attacks on deep learning models trained for 3D neuroimaging tasks in a centralized as well as decentralized setup. We demonstrate feasible attacks on brain age prediction models (deep learning models that predict a person's age from their brain MRI scan). We correctly identified whether an MRI scan was used in model training with a 60% to over 80% success rate depending on model complexity and security assumptions.
Improved Brain Age Estimation with Slice-based Set Networks
Feb 09, 2021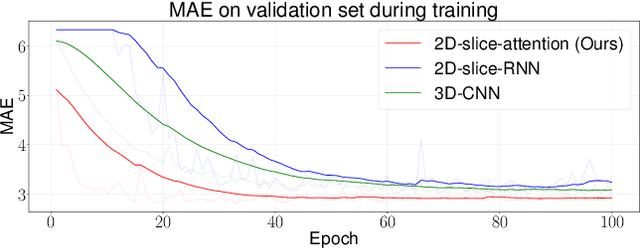
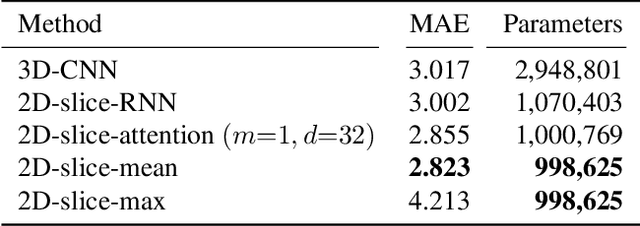
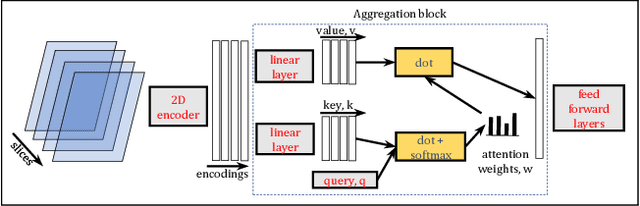
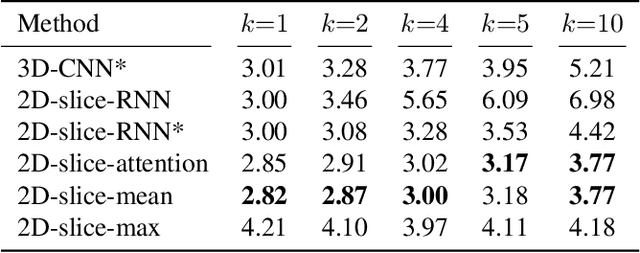
Abstract:Deep Learning for neuroimaging data is a promising but challenging direction. The high dimensionality of 3D MRI scans makes this endeavor compute and data-intensive. Most conventional 3D neuroimaging methods use 3D-CNN-based architectures with a large number of parameters and require more time and data to train. Recently, 2D-slice-based models have received increasing attention as they have fewer parameters and may require fewer samples to achieve comparable performance. In this paper, we propose a new architecture for BrainAGE prediction. The proposed architecture works by encoding each 2D slice in an MRI with a deep 2D-CNN model. Next, it combines the information from these 2D-slice encodings using set networks or permutation invariant layers. Experiments on the BrainAGE prediction problem, using the UK Biobank dataset, showed that the model with the permutation invariant layers trains faster and provides better predictions compared to other state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge