Chantal M. W. Tax
aDWI-BIDS: an extension to the brain imaging data structure for advanced diffusion weighted imaging
Apr 12, 2021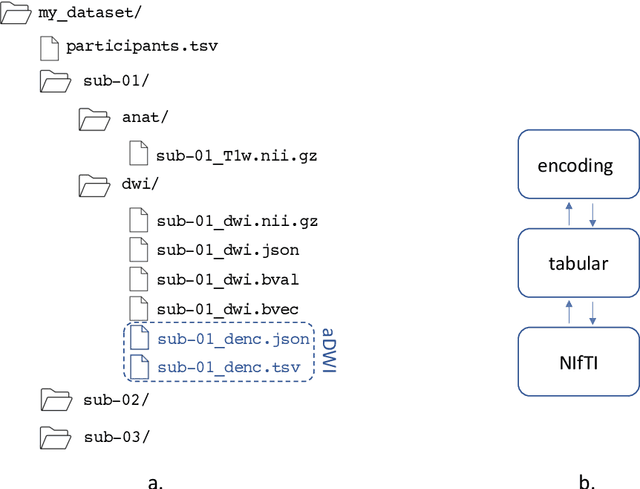
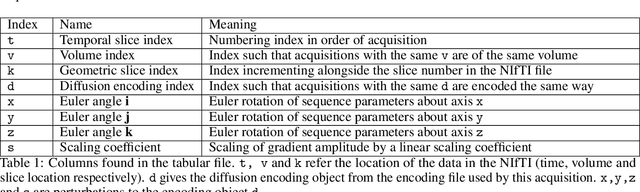
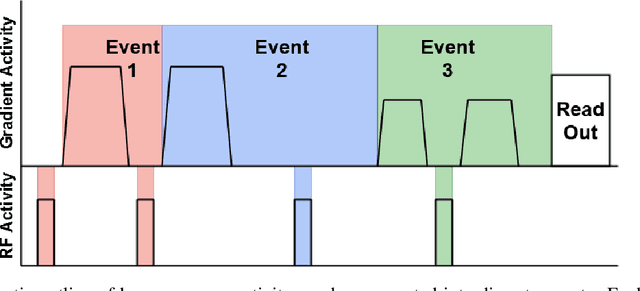
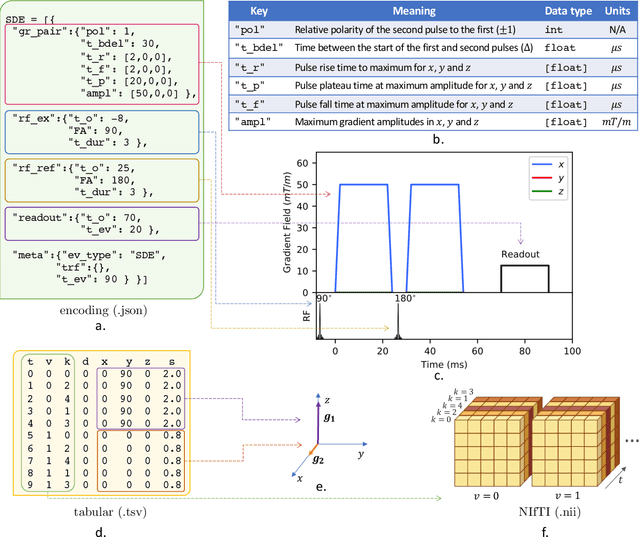
Abstract:Diffusion weighted imaging techniques permit us to infer microstructural detail in biological tissue in vivo and noninvasively. Modern sequences are based on advanced diffusion encoding schemes, allowing probing of more revealing measures of tissue microstructure than the standard apparent diffusion coefficient or fractional anisotropy. Though these methods may result in faster or more revealing acquisitions, they generally demand prior knowledge of sequence-specific parameters for which there is no accepted sharing standard. Here, we present a metadata labelling scheme suitable for the needs of developers and users within the diffusion neuroimaging community alike: a lightweight, unambiguous parametric map relaying acqusition parameters. This extensible scheme supports a wide spectrum of diffusion encoding methods, from single diffusion encoding to highly complex sequences involving arbitrary gradient waveforms. Built under the brain imaging data structure (BIDS), it allows storage of advanced diffusion MRI data comprehensively alongside any other neuroimaging information, facilitating processing pipelines and multimodal analyses. We illustrate the usefulness of this BIDS-extension with a range of example data, and discuss the extension's impact on pre- and post-processing software.
Tractometry-based Anomaly Detection for Single-subject White Matter Analysis
May 25, 2020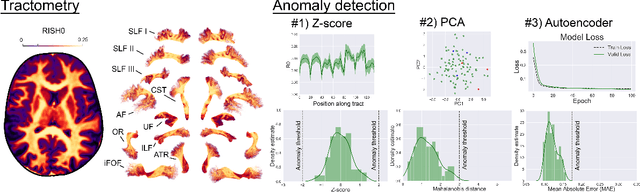
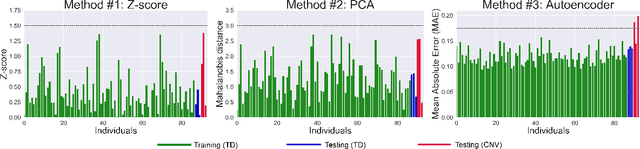
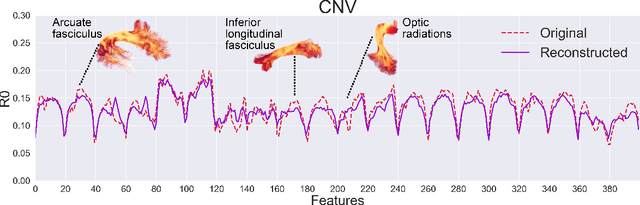
Abstract:There is an urgent need for a paradigm shift from group-wise comparisons to individual diagnosis in diffusion MRI (dMRI) to enable the analysis of rare cases and clinically-heterogeneous groups. Deep autoencoders have shown great potential to detect anomalies in neuroimaging data. We present a framework that operates on the manifold of white matter (WM) pathways to learn normative microstructural features, and discriminate those at genetic risk from controls in a paediatric population.
* Medical Imaging with Deep Learning (MIDL2020) Conference Short Paper
Multi-Stage Prediction Networks for Data Harmonization
Jul 26, 2019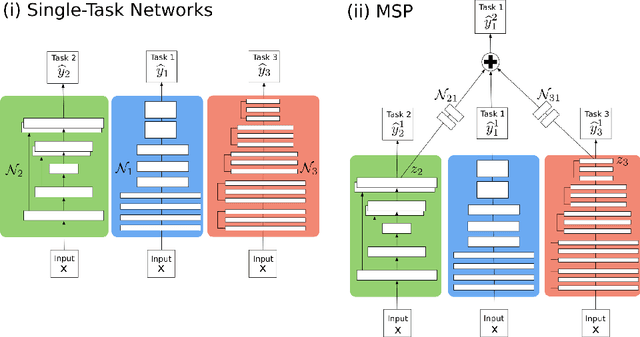
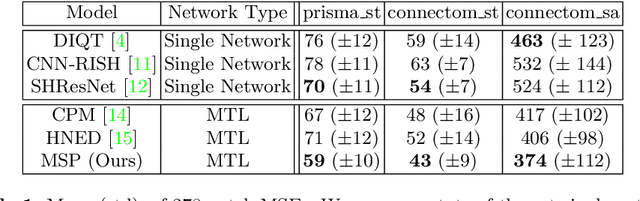
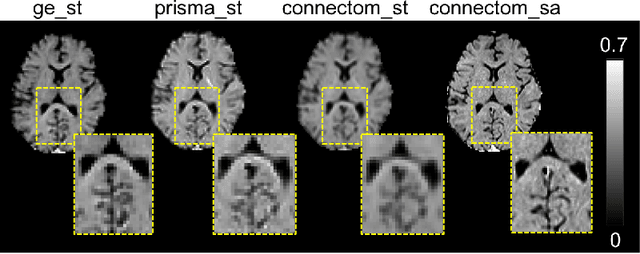
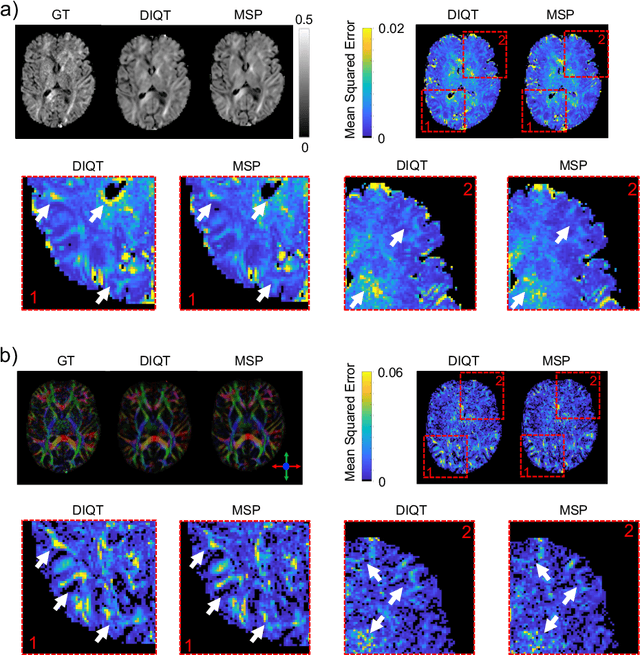
Abstract:In this paper, we introduce multi-task learning (MTL) to data harmonization (DH); where we aim to harmonize images across different acquisition platforms and sites. This allows us to integrate information from multiple acquisitions and improve the predictive performance and learning efficiency of the harmonization model. Specifically, we introduce the Multi Stage Prediction (MSP) Network, a MTL framework that incorporates neural networks of potentially disparate architectures, trained for different individual acquisition platforms, into a larger architecture that is refined in unison. The MSP utilizes high-level features of single networks for individual tasks, as inputs of additional neural networks to inform the final prediction, therefore exploiting redundancy across tasks to make the most of limited training data. We validate our methods on a dMRI harmonization challenge dataset, where we predict three modern platform types, from one obtained from an old scanner. We show how MTL architectures, such as the MSP, produce around 20\% improvement of patch-based mean-squared error over current state-of-the-art methods and that our MSP outperforms off-the-shelf MTL networks. Our code is available https://github.com/sbb-gh/ .
Scanner Invariant Representations for Diffusion MRI Harmonization
Apr 10, 2019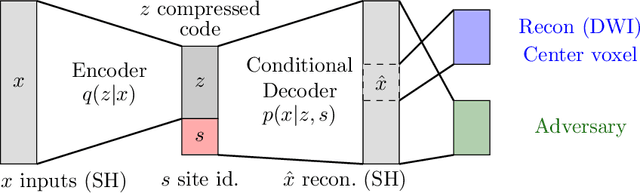
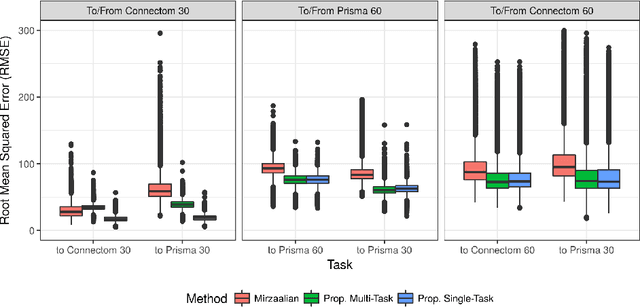
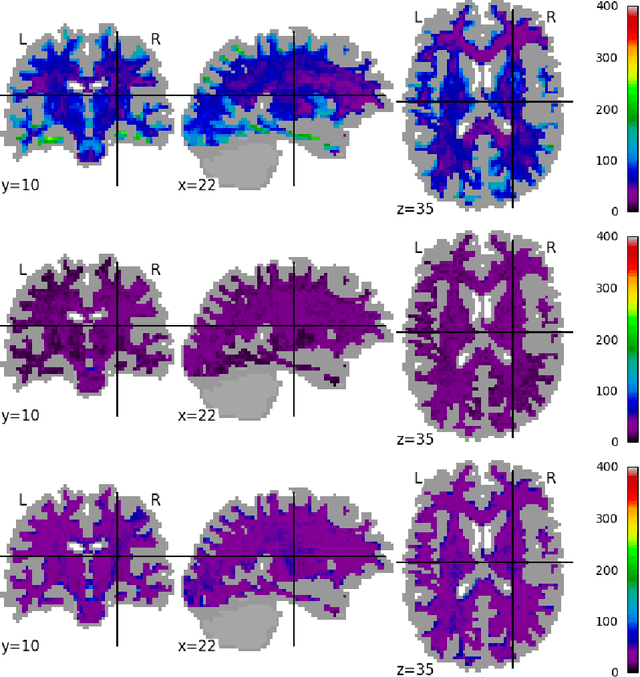
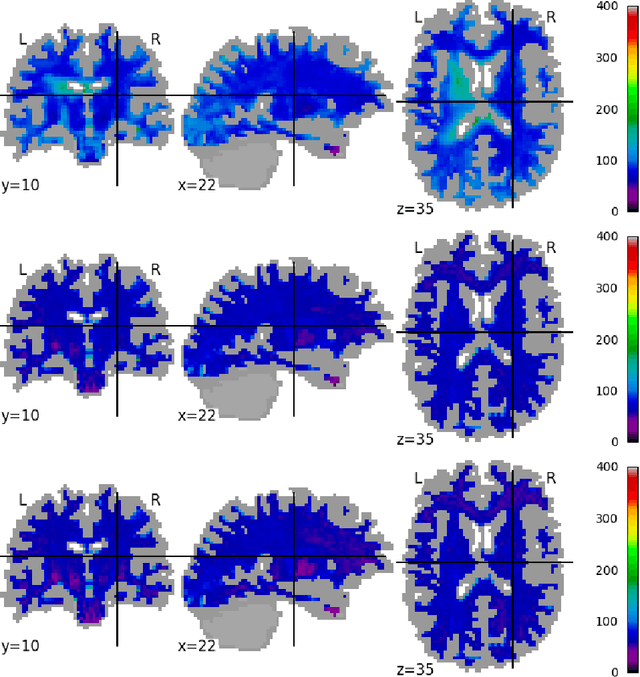
Abstract:Pooled imaging data from multiple sources is subject to variation between the sources. Correcting for these biases has become incredibly important as the size of imaging studies increases and the multi-site case becomes more common. We propose learning an intermediate representation invariant to site/protocol variables, a technique adapted from information theory-based algorithmic fairness; by leveraging the data processing inequality, such a representation can then be used to create an image reconstruction that is uninformative of its original source, yet still faithful to the underlying structure. To implement this, we use a machine learning method based on variational auto-encoders (VAE) to construct scanner invariant encodings of the imaging data. To evaluate our method, we use training data from the 2018 CDMRI Challenge Harmonization dataset. Our proposed method shows improvements on independent test data relative to a recently published baseline method.
Spherical Harmonic Residual Network for Diffusion Signal Harmonization
Aug 05, 2018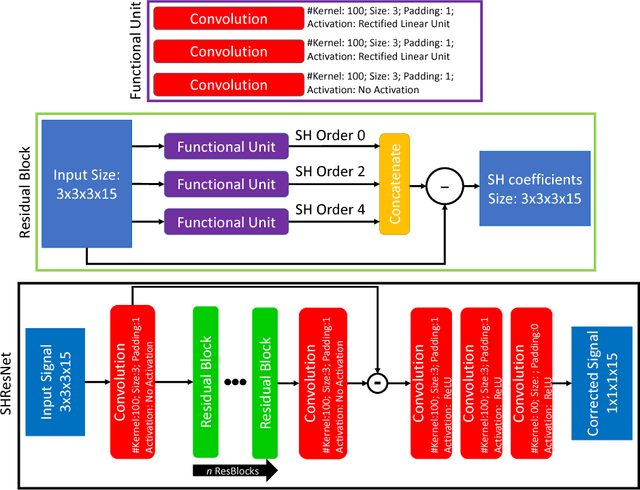

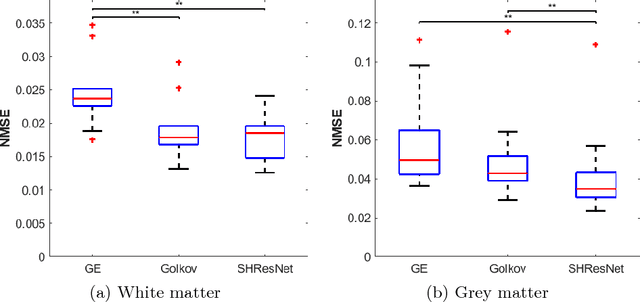
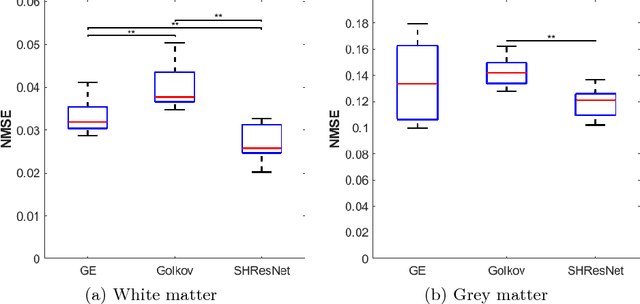
Abstract:Diffusion imaging is an important method in the field of neuroscience, as it is sensitive to changes within the tissue microstructure of the human brain. However, a major challenge when using MRI to derive quantitative measures is that the use of different scanners, as used in multi-site group studies, introduces measurement variability. This can lead to an increased variance in quantitative metrics, even if the same brain is scanned. Contrary to the assumption that these characteristics are comparable and similar, small changes in these values are observed in many clinical studies, hence harmonization of the signals is essential. In this paper, we present a method that does not require additional preprocessing, such as segmentation or registration, and harmonizes the signal based on a deep learning residual network. For this purpose, a training database is required, which consist of the same subjects, scanned on different scanners. The results show that harmonized signals are significantly more similar to the ground truth signal compared to no harmonization, but also improve in comparison to another deep learning method. The same effect is also demonstrated in commonly used metrics derived from the diffusion MRI signal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge