Paul E. Kinahan
Scale-aware Adaptive Supervised Network with Limited Medical Annotations
Jan 02, 2026Abstract:Medical image segmentation faces critical challenges in semi-supervised learning scenarios due to severe annotation scarcity requiring expert radiological knowledge, significant inter-annotator variability across different viewpoints and expertise levels, and inadequate multi-scale feature integration for precise boundary delineation in complex anatomical structures. Existing semi-supervised methods demonstrate substantial performance degradation compared to fully supervised approaches, particularly in small target segmentation and boundary refinement tasks. To address these fundamental challenges, we propose SASNet (Scale-aware Adaptive Supervised Network), a dual-branch architecture that leverages both low-level and high-level feature representations through novel scale-aware adaptive reweight mechanisms. Our approach introduces three key methodological innovations, including the Scale-aware Adaptive Reweight strategy that dynamically weights pixel-wise predictions using temporal confidence accumulation, the View Variance Enhancement mechanism employing 3D Fourier domain transformations to simulate annotation variability, and segmentation-regression consistency learning through signed distance map algorithms for enhanced boundary precision. These innovations collectively address the core limitations of existing semi-supervised approaches by integrating spatial, temporal, and geometric consistency principles within a unified optimization framework. Comprehensive evaluation across LA, Pancreas-CT, and BraTS datasets demonstrates that SASNet achieves superior performance with limited labeled data, surpassing state-of-the-art semi-supervised methods while approaching fully supervised performance levels. The source code for SASNet is available at https://github.com/HUANGLIZI/SASNet.
VisionUnite: A Vision-Language Foundation Model for Ophthalmology Enhanced with Clinical Knowledge
Aug 05, 2024



Abstract:The need for improved diagnostic methods in ophthalmology is acute, especially in the less developed regions with limited access to specialists and advanced equipment. Therefore, we introduce VisionUnite, a novel vision-language foundation model for ophthalmology enhanced with clinical knowledge. VisionUnite has been pretrained on an extensive dataset comprising 1.24 million image-text pairs, and further refined using our proposed MMFundus dataset, which includes 296,379 high-quality fundus image-text pairs and 889,137 simulated doctor-patient dialogue instances. Our experiments indicate that VisionUnite outperforms existing generative foundation models such as GPT-4V and Gemini Pro. It also demonstrates diagnostic capabilities comparable to junior ophthalmologists. VisionUnite performs well in various clinical scenarios including open-ended multi-disease diagnosis, clinical explanation, and patient interaction, making it a highly versatile tool for initial ophthalmic disease screening. VisionUnite can also serve as an educational aid for junior ophthalmologists, accelerating their acquisition of knowledge regarding both common and rare ophthalmic conditions. VisionUnite represents a significant advancement in ophthalmology, with broad implications for diagnostics, medical education, and understanding of disease mechanisms.
Issues and Challenges in Applications of Artificial Intelligence to Nuclear Medicine -- The Bethesda Report (AI Summit 2022)
Nov 07, 2022

Abstract:The SNMMI Artificial Intelligence (SNMMI-AI) Summit, organized by the SNMMI AI Task Force, took place in Bethesda, MD on March 21-22, 2022. It brought together various community members and stakeholders from academia, healthcare, industry, patient representatives, and government (NIH, FDA), and considered various key themes to envision and facilitate a bright future for routine, trustworthy use of AI in nuclear medicine. In what follows, essential issues, challenges, controversies and findings emphasized in the meeting are summarized.
Synthetic PET via Domain Translation of 3D MRI
Jun 11, 2022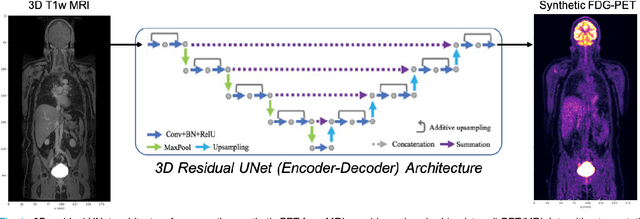
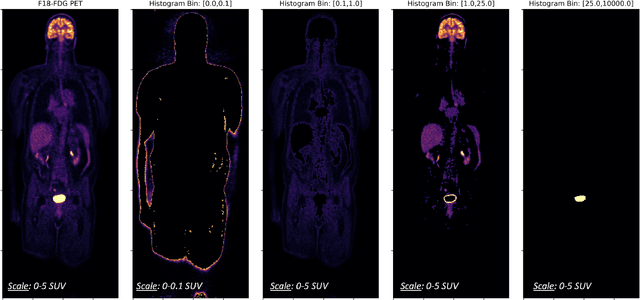
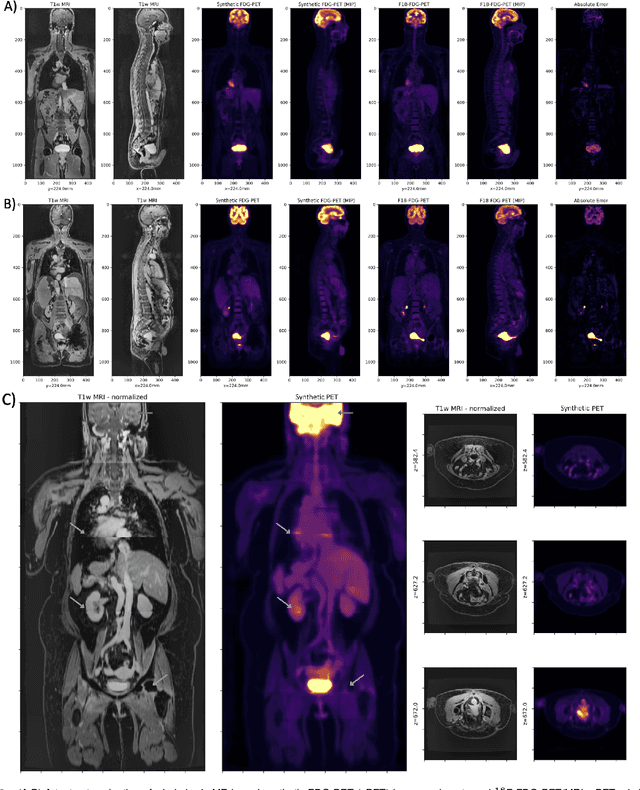
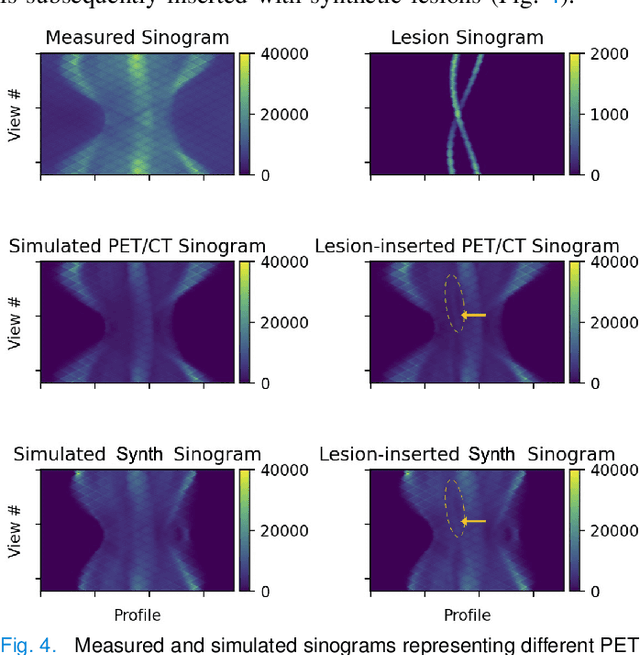
Abstract:Historically, patient datasets have been used to develop and validate various reconstruction algorithms for PET/MRI and PET/CT. To enable such algorithm development, without the need for acquiring hundreds of patient exams, in this paper we demonstrate a deep learning technique to generate synthetic but realistic whole-body PET sinograms from abundantly-available whole-body MRI. Specifically, we use a dataset of 56 $^{18}$F-FDG-PET/MRI exams to train a 3D residual UNet to predict physiologic PET uptake from whole-body T1-weighted MRI. In training we implemented a balanced loss function to generate realistic uptake across a large dynamic range and computed losses along tomographic lines of response to mimic the PET acquisition. The predicted PET images are forward projected to produce synthetic PET time-of-flight (ToF) sinograms that can be used with vendor-provided PET reconstruction algorithms, including using CT-based attenuation correction (CTAC) and MR-based attenuation correction (MRAC). The resulting synthetic data recapitulates physiologic $^{18}$F-FDG uptake, e.g. high uptake localized to the brain and bladder, as well as uptake in liver, kidneys, heart and muscle. To simulate abnormalities with high uptake, we also insert synthetic lesions. We demonstrate that this synthetic PET data can be used interchangeably with real PET data for the PET quantification task of comparing CT and MR-based attenuation correction methods, achieving $\leq 7.6\%$ error in mean-SUV compared to using real data. These results together show that the proposed synthetic PET data pipeline can be reasonably used for development, evaluation, and validation of PET/MRI reconstruction methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge