Mehmet Yigit Avci
deepc GmbH, Blumenstrasse 28, 80331 Munich, Germany
DIST-CLIP: Arbitrary Metadata and Image Guided MRI Harmonization via Disentangled Anatomy-Contrast Representations
Dec 08, 2025Abstract:Deep learning holds immense promise for transforming medical image analysis, yet its clinical generalization remains profoundly limited. A major barrier is data heterogeneity. This is particularly true in Magnetic Resonance Imaging, where scanner hardware differences, diverse acquisition protocols, and varying sequence parameters introduce substantial domain shifts that obscure underlying biological signals. Data harmonization methods aim to reduce these instrumental and acquisition variability, but existing approaches remain insufficient. When applied to imaging data, image-based harmonization approaches are often restricted by the need for target images, while existing text-guided methods rely on simplistic labels that fail to capture complex acquisition details or are typically restricted to datasets with limited variability, failing to capture the heterogeneity of real-world clinical environments. To address these limitations, we propose DIST-CLIP (Disentangled Style Transfer with CLIP Guidance), a unified framework for MRI harmonization that flexibly uses either target images or DICOM metadata for guidance. Our framework explicitly disentangles anatomical content from image contrast, with the contrast representations being extracted using pre-trained CLIP encoders. These contrast embeddings are then integrated into the anatomical content via a novel Adaptive Style Transfer module. We trained and evaluated DIST-CLIP on diverse real-world clinical datasets, and showed significant improvements in performance when compared against state-of-the-art methods in both style translation fidelity and anatomical preservation, offering a flexible solution for style transfer and standardizing MRI data. Our code and weights will be made publicly available upon publication.
Unsupervised Analysis of Alzheimer's Disease Signatures using 3D Deformable Autoencoders
Jul 04, 2024



Abstract:With the increasing incidence of neurodegenerative diseases such as Alzheimer's Disease (AD), there is a need for further research that enhances detection and monitoring of the diseases. We present MORPHADE (Morphological Autoencoders for Alzheimer's Disease Detection), a novel unsupervised learning approach which uses deformations to allow the analysis of 3D T1-weighted brain images. To the best of our knowledge, this is the first use of deformations with deep unsupervised learning to not only detect, but also localize and assess the severity of structural changes in the brain due to AD. We obtain markedly higher anomaly scores in clinically important areas of the brain in subjects with AD compared to healthy controls, showcasing that our method is able to effectively locate AD-related atrophy. We additionally observe a visual correlation between the severity of atrophy highlighted in our anomaly maps and medial temporal lobe atrophy scores evaluated by a clinical expert. Finally, our method achieves an AUROC of 0.80 in detecting AD, out-performing several supervised and unsupervised baselines. We believe our framework shows promise as a tool towards improved understanding, monitoring and detection of AD. To support further research and application, we have made our code publicly available at github.com/ci-ber/MORPHADE.
Simulation of acquisition shifts in T2 Flair MR images to stress test AI segmentation networks
Nov 03, 2023



Abstract:Purpose: To provide a simulation framework for routine neuroimaging test data, which allows for "stress testing" of deep segmentation networks against acquisition shifts that commonly occur in clinical practice for T2 weighted (T2w) fluid attenuated inversion recovery (FLAIR) Magnetic Resonance Imaging (MRI) protocols. Approach: The approach simulates "acquisition shift derivatives" of MR images based on MR signal equations. Experiments comprise the validation of the simulated images by real MR scans and example stress tests on state-of-the-art MS lesion segmentation networks to explore a generic model function to describe the F1 score in dependence of the contrast-affecting sequence parameters echo time (TE) and inversion time (TI). Results: The differences between real and simulated images range up to 19 % in gray and white matter for extreme parameter settings. For the segmentation networks under test the F1 score dependency on TE and TI can be well described by quadratic model functions (R^2 > 0.9). The coefficients of the model functions indicate that changes of TE have more influence on the model performance than TI. Conclusions: We show that these deviations are in the range of values as may be caused by erroneous or individual differences of relaxation times as described by literature. The coefficients of the F1 model function allow for quantitative comparison of the influences of TE and TI. Limitations arise mainly from tissues with the low baseline signal (like CSF) and when the protocol contains contrast-affecting measures that cannot be modelled due to missing information in the DICOM header.
Quantifying the uncertainty of neural networks using Monte Carlo dropout for deep learning based quantitative MRI
Dec 02, 2021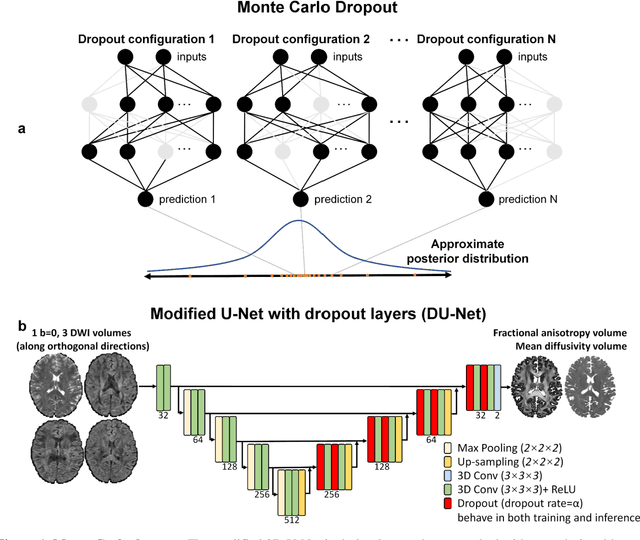
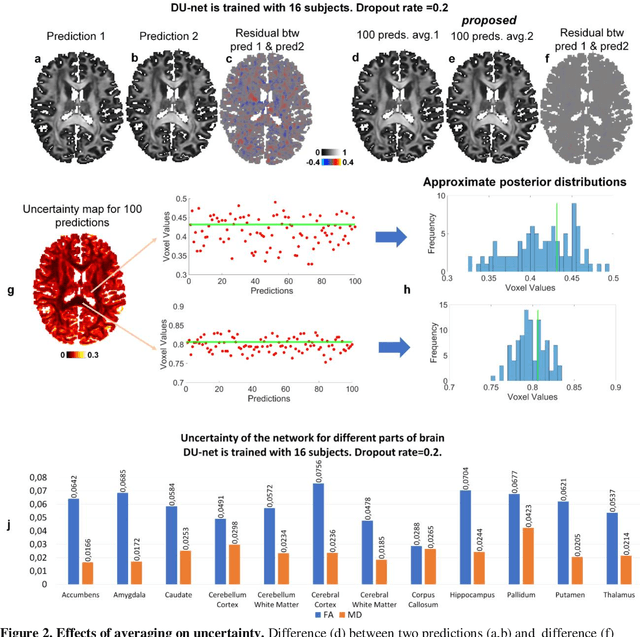

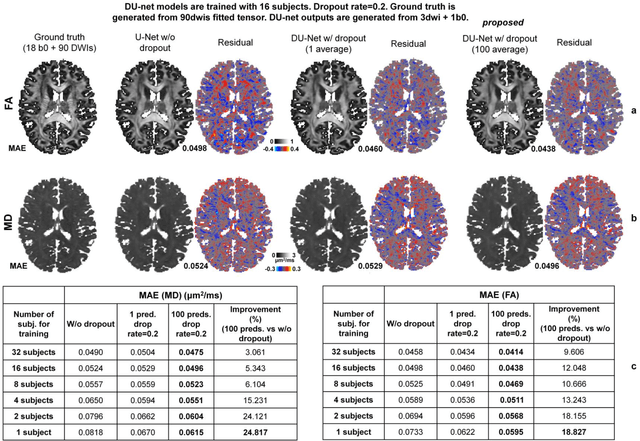
Abstract:Dropout is conventionally used during the training phase as regularization method and for quantifying uncertainty in deep learning. We propose to use dropout during training as well as inference steps, and average multiple predictions to improve the accuracy, while reducing and quantifying the uncertainty. The results are evaluated for fractional anisotropy (FA) and mean diffusivity (MD) maps which are obtained from only 3 direction scans. With our method, accuracy can be improved significantly compared to network outputs without dropout, especially when the training dataset is small. Moreover, confidence maps are generated which may aid in diagnosis of unseen pathology or artifacts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge