Mehmet Yigitsoy
deepc GmbH, Blumenstrasse 28, 80331 Munich, Germany
DIST-CLIP: Arbitrary Metadata and Image Guided MRI Harmonization via Disentangled Anatomy-Contrast Representations
Dec 08, 2025Abstract:Deep learning holds immense promise for transforming medical image analysis, yet its clinical generalization remains profoundly limited. A major barrier is data heterogeneity. This is particularly true in Magnetic Resonance Imaging, where scanner hardware differences, diverse acquisition protocols, and varying sequence parameters introduce substantial domain shifts that obscure underlying biological signals. Data harmonization methods aim to reduce these instrumental and acquisition variability, but existing approaches remain insufficient. When applied to imaging data, image-based harmonization approaches are often restricted by the need for target images, while existing text-guided methods rely on simplistic labels that fail to capture complex acquisition details or are typically restricted to datasets with limited variability, failing to capture the heterogeneity of real-world clinical environments. To address these limitations, we propose DIST-CLIP (Disentangled Style Transfer with CLIP Guidance), a unified framework for MRI harmonization that flexibly uses either target images or DICOM metadata for guidance. Our framework explicitly disentangles anatomical content from image contrast, with the contrast representations being extracted using pre-trained CLIP encoders. These contrast embeddings are then integrated into the anatomical content via a novel Adaptive Style Transfer module. We trained and evaluated DIST-CLIP on diverse real-world clinical datasets, and showed significant improvements in performance when compared against state-of-the-art methods in both style translation fidelity and anatomical preservation, offering a flexible solution for style transfer and standardizing MRI data. Our code and weights will be made publicly available upon publication.
Differentially Private Active Learning: Balancing Effective Data Selection and Privacy
Oct 01, 2024



Abstract:Active learning (AL) is a widely used technique for optimizing data labeling in machine learning by iteratively selecting, labeling, and training on the most informative data. However, its integration with formal privacy-preserving methods, particularly differential privacy (DP), remains largely underexplored. While some works have explored differentially private AL for specialized scenarios like online learning, the fundamental challenge of combining AL with DP in standard learning settings has remained unaddressed, severely limiting AL's applicability in privacy-sensitive domains. This work addresses this gap by introducing differentially private active learning (DP-AL) for standard learning settings. We demonstrate that naively integrating DP-SGD training into AL presents substantial challenges in privacy budget allocation and data utilization. To overcome these challenges, we propose step amplification, which leverages individual sampling probabilities in batch creation to maximize data point participation in training steps, thus optimizing data utilization. Additionally, we investigate the effectiveness of various acquisition functions for data selection under privacy constraints, revealing that many commonly used functions become impractical. Our experiments on vision and natural language processing tasks show that DP-AL can improve performance for specific datasets and model architectures. However, our findings also highlight the limitations of AL in privacy-constrained environments, emphasizing the trade-offs between privacy, model accuracy, and data selection accuracy.
Simulation of acquisition shifts in T2 Flair MR images to stress test AI segmentation networks
Nov 03, 2023



Abstract:Purpose: To provide a simulation framework for routine neuroimaging test data, which allows for "stress testing" of deep segmentation networks against acquisition shifts that commonly occur in clinical practice for T2 weighted (T2w) fluid attenuated inversion recovery (FLAIR) Magnetic Resonance Imaging (MRI) protocols. Approach: The approach simulates "acquisition shift derivatives" of MR images based on MR signal equations. Experiments comprise the validation of the simulated images by real MR scans and example stress tests on state-of-the-art MS lesion segmentation networks to explore a generic model function to describe the F1 score in dependence of the contrast-affecting sequence parameters echo time (TE) and inversion time (TI). Results: The differences between real and simulated images range up to 19 % in gray and white matter for extreme parameter settings. For the segmentation networks under test the F1 score dependency on TE and TI can be well described by quadratic model functions (R^2 > 0.9). The coefficients of the model functions indicate that changes of TE have more influence on the model performance than TI. Conclusions: We show that these deviations are in the range of values as may be caused by erroneous or individual differences of relaxation times as described by literature. The coefficients of the F1 model function allow for quantitative comparison of the influences of TE and TI. Limitations arise mainly from tissues with the low baseline signal (like CSF) and when the protocol contains contrast-affecting measures that cannot be modelled due to missing information in the DICOM header.
Deep Attention Based Semi-Supervised 2D-Pose Estimation for Surgical Instruments
Dec 10, 2019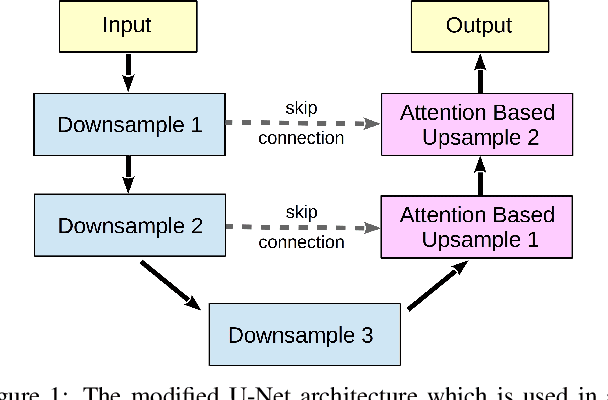
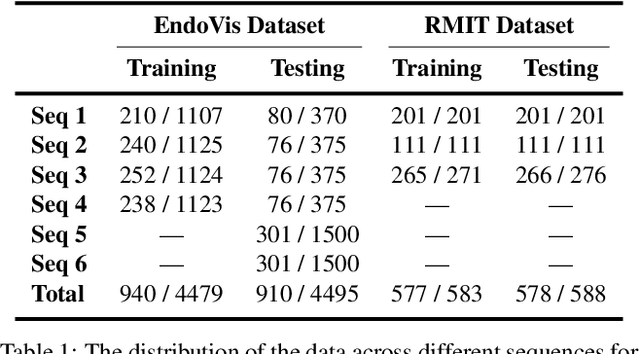
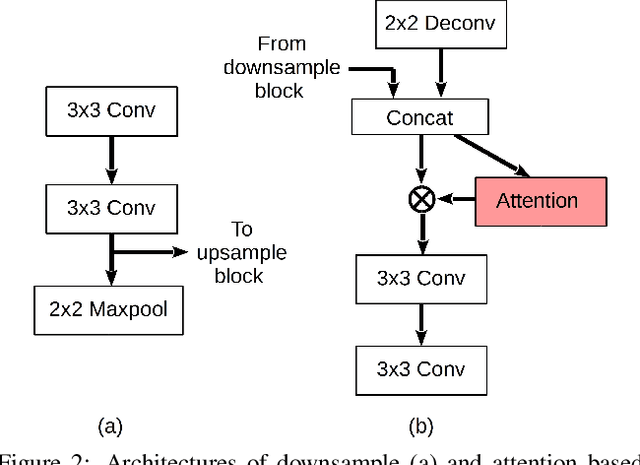
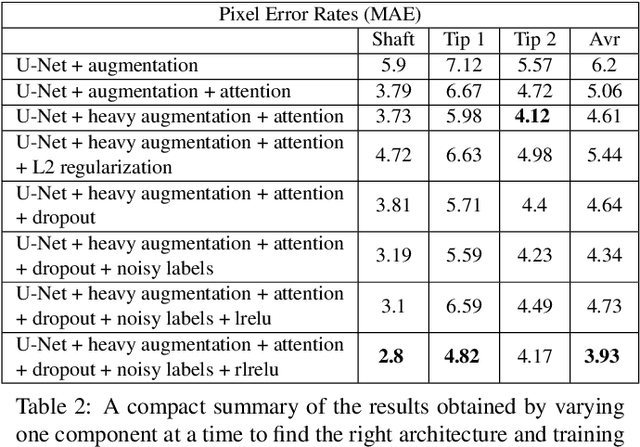
Abstract:For many practical problems and applications, it is not feasible to create a vast and accurately labeled dataset, which restricts the application of deep learning in many areas. Semi-supervised learning algorithms intend to improve performance by also leveraging unlabeled data. This is very valuable for 2D-pose estimation task where data labeling requires substantial time and is subject to noise. This work aims to investigate if semi-supervised learning techniques can achieve acceptable performance level that makes using these algorithms during training justifiable. To this end, a lightweight network architecture is introduced and mean teacher, virtual adversarial training and pseudo-labeling algorithms are evaluated on 2D-pose estimation for surgical instruments. For the applicability of pseudo-labelling algorithm, we propose a novel confidence measure, total variation. Experimental results show that utilization of semi-supervised learning improves the performance on unseen geometries drastically while maintaining high accuracy for seen geometries. For RMIT benchmark, our lightweight architecture outperforms state-of-the-art with supervised learning. For Endovis benchmark, pseudo-labelling algorithm improves the supervised baseline achieving the new state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge