Keyi Li
Towards Artificial Intelligence Research Assistant for Expert-Involved Learning
May 03, 2025Abstract:Large Language Models (LLMs) and Large Multi-Modal Models (LMMs) have emerged as transformative tools in scientific research, yet their reliability and specific contributions to biomedical applications remain insufficiently characterized. In this study, we present \textbf{AR}tificial \textbf{I}ntelligence research assistant for \textbf{E}xpert-involved \textbf{L}earning (ARIEL), a multimodal dataset designed to benchmark and enhance two critical capabilities of LLMs and LMMs in biomedical research: summarizing extensive scientific texts and interpreting complex biomedical figures. To facilitate rigorous assessment, we create two open-source sets comprising biomedical articles and figures with designed questions. We systematically benchmark both open- and closed-source foundation models, incorporating expert-driven human evaluations conducted by doctoral-level experts. Furthermore, we improve model performance through targeted prompt engineering and fine-tuning strategies for summarizing research papers, and apply test-time computational scaling to enhance the reasoning capabilities of LMMs, achieving superior accuracy compared to human-expert corrections. We also explore the potential of using LMM Agents to generate scientific hypotheses from diverse multimodal inputs. Overall, our results delineate clear strengths and highlight significant limitations of current foundation models, providing actionable insights and guiding future advancements in deploying large-scale language and multi-modal models within biomedical research.
Every Component Counts: Rethinking the Measure of Success for Medical Semantic Segmentation in Multi-Instance Segmentation Tasks
Oct 24, 2024



Abstract:We present Connected-Component~(CC)-Metrics, a novel semantic segmentation evaluation protocol, targeted to align existing semantic segmentation metrics to a multi-instance detection scenario in which each connected component matters. We motivate this setup in the common medical scenario of semantic metastases segmentation in a full-body PET/CT. We show how existing semantic segmentation metrics suffer from a bias towards larger connected components contradicting the clinical assessment of scans in which tumor size and clinical relevance are uncorrelated. To rebalance existing segmentation metrics, we propose to evaluate them on a per-component basis thus giving each tumor the same weight irrespective of its size. To match predictions to ground-truth segments, we employ a proximity-based matching criterion, evaluating common metrics locally at the component of interest. Using this approach, we break free of biases introduced by large metastasis for overlap-based metrics such as Dice or Surface Dice. CC-Metrics also improves distance-based metrics such as Hausdorff Distances which are uninformative for small changes that do not influence the maximum or 95th percentile, and avoids pitfalls introduced by directly combining counting-based metrics with overlap-based metrics as it is done in Panoptic Quality.
MaskMatch: Boosting Semi-Supervised Learning Through Mask Autoencoder-Driven Feature Learning
May 10, 2024



Abstract:Conventional methods in semi-supervised learning (SSL) often face challenges related to limited data utilization, mainly due to their reliance on threshold-based techniques for selecting high-confidence unlabeled data during training. Various efforts (e.g., FreeMatch) have been made to enhance data utilization by tweaking the thresholds, yet none have managed to use 100% of the available data. To overcome this limitation and improve SSL performance, we introduce \algo, a novel algorithm that fully utilizes unlabeled data to boost semi-supervised learning. \algo integrates a self-supervised learning strategy, i.e., Masked Autoencoder (MAE), that uses all available data to enforce the visual representation learning. This enables the SSL algorithm to leverage all available data, including samples typically filtered out by traditional methods. In addition, we propose a synthetic data training approach to further increase data utilization and improve generalization. These innovations lead \algo to achieve state-of-the-art results on challenging datasets. For instance, on CIFAR-100 with 2 labels per class, STL-10 with 4 labels per class, and Euro-SAT with 2 labels per class, \algo achieves low error rates of 18.71%, 9.47%, and 3.07%, respectively. The code will be made publicly available.
Exploring Runtime Decision Support for Trauma Resuscitation
Jul 06, 2022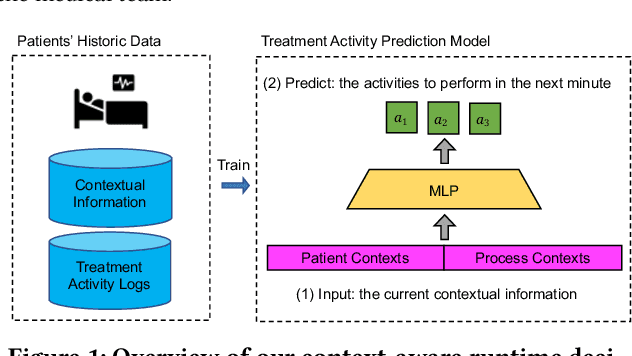
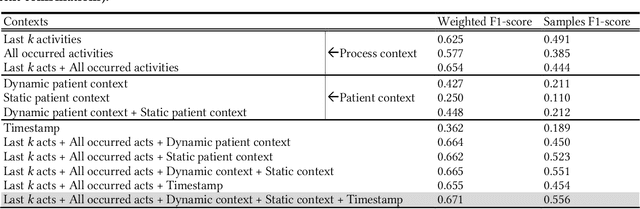
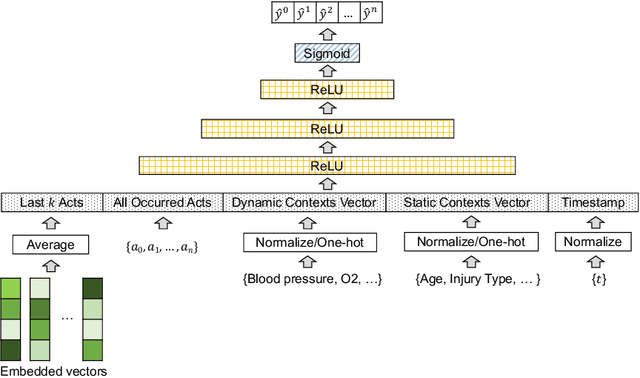
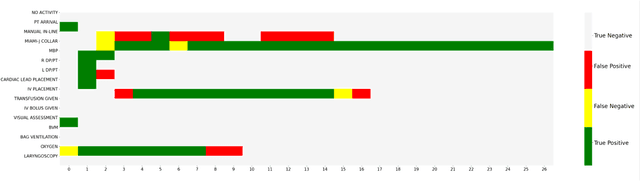
Abstract:AI-based recommender systems have been successfully applied in many domains (e.g., e-commerce, feeds ranking). Medical experts believe that incorporating such methods into a clinical decision support system may help reduce medical team errors and improve patient outcomes during treatment processes (e.g., trauma resuscitation, surgical processes). Limited research, however, has been done to develop automatic data-driven treatment decision support. We explored the feasibility of building a treatment recommender system to provide runtime next-minute activity predictions. The system uses patient context (e.g., demographics and vital signs) and process context (e.g., activities) to continuously predict activities that will be performed in the next minute. We evaluated our system on a pre-recorded dataset of trauma resuscitation and conducted an ablation study on different model variants. The best model achieved an average F1-score of 0.67 for 61 activity types. We include medical team feedback and discuss the future work.
Generating Privacy-Preserving Process Data with Deep Generative Models
Mar 15, 2022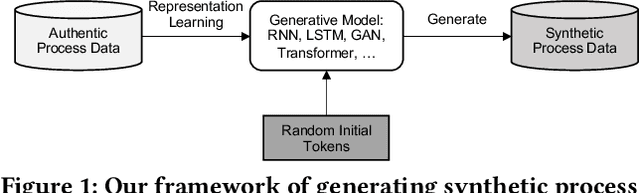
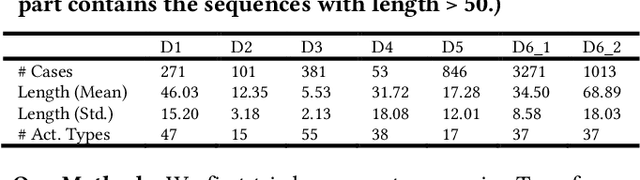
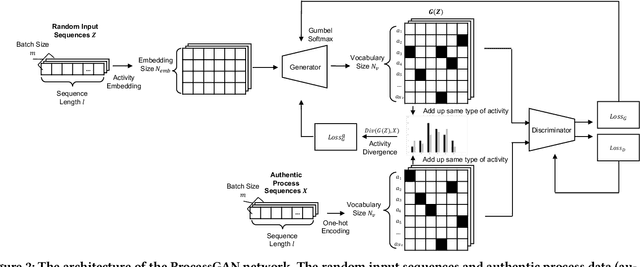
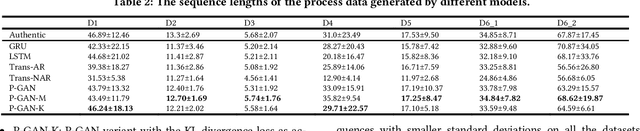
Abstract:Process data with confidential information cannot be shared directly in public, which hinders the research in process data mining and analytics. Data encryption methods have been studied to protect the data, but they still may be decrypted, which leads to individual identification. We experimented with different models of representation learning and used the learned model to generate synthetic process data. We introduced an adversarial generative network for process data generation (ProcessGAN) with two Transformer networks for the generator and the discriminator. We evaluated ProcessGAN and traditional models on six real-world datasets, of which two are public and four are collected in medical domains. We used statistical metrics and supervised learning scores to evaluate the synthetic data. We also used process mining to discover workflows for the authentic and synthetic datasets and had medical experts evaluate the clinical applicability of the synthetic workflows. We found that ProcessGAN outperformed traditional sequential models when trained on small authentic datasets of complex processes. ProcessGAN better represented the long-range dependencies between the activities, which is important for complicated processes such as the medical processes. Traditional sequential models performed better when trained on large data of simple processes. We conclude that ProcessGAN can generate a large amount of sharable synthetic process data indistinguishable from authentic data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge