Kai Ma
School of Electrical Engineering, Yanshan University, Qinhuangdao, China
Uncertainty-Guided Domain Alignment for Layer Segmentation in OCT Images
Aug 30, 2019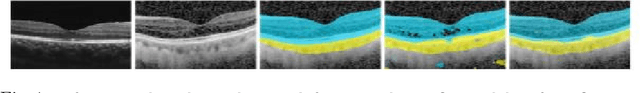
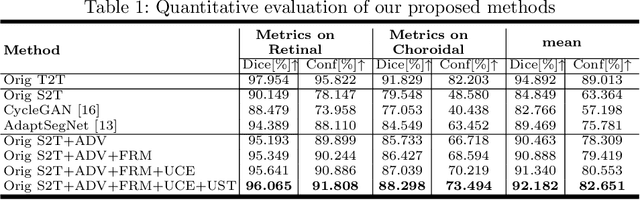
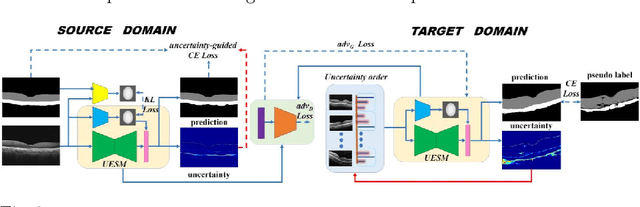
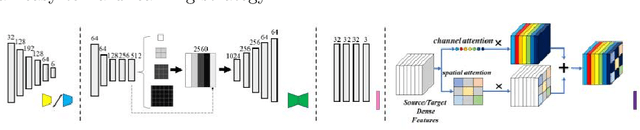
Abstract:Automatic and accurate segmentation for retinal and choroidal layers of Optical Coherence Tomography (OCT) is crucial for detection of various ocular diseases. However, because of the variations in different equipments, OCT data obtained from different manufacturers might encounter appearance discrepancy, which could lead to performance fluctuation to a deep neural network. In this paper, we propose an uncertainty-guided domain alignment method to aim at alleviating this problem to transfer discriminative knowledge across distinct domains. We disign a novel uncertainty-guided cross-entropy loss for boosting the performance over areas with high uncertainty. An uncertainty-guided curriculum transfer strategy is developed for the self-training (ST), which regards uncertainty as efficient and effective guidance to optimize the learning process in target domain. Adversarial learning with feature recalibration module (FRM) is applied to transfer informative knowledge from the domain feature spaces adaptively. The experiments on two OCT datasets show that the proposed methods can obtain significant segmentation improvements compared with the baseline models.
Semi-supervised Breast Lesion Detection in Ultrasound Video Based on Temporal Coherence
Jul 16, 2019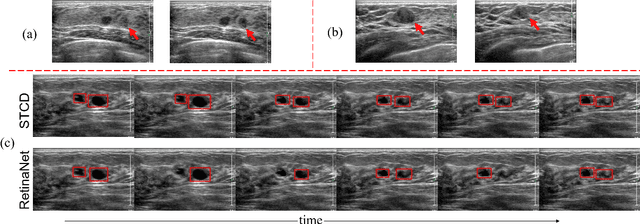

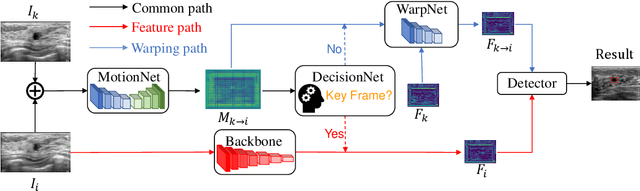
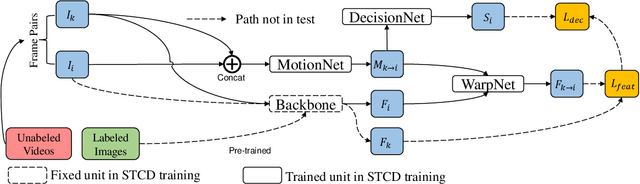
Abstract:Breast lesion detection in ultrasound video is critical for computer-aided diagnosis. However, detecting lesion in video is quite challenging due to the blurred lesion boundary, high similarity to soft tissue and lack of video annotations. In this paper, we propose a semi-supervised breast lesion detection method based on temporal coherence which can detect the lesion more accurately. We aggregate features extracted from the historical key frames with adaptive key-frame scheduling strategy. Our proposed method accomplishes the unlabeled videos detection task by leveraging the supervision information from a different set of labeled images. In addition, a new WarpNet is designed to replace both the traditional spatial warping and feature aggregation operation, leading to a tremendous increase in speed. Experiments on 1,060 2D ultrasound sequences demonstrate that our proposed method achieves state-of-the-art video detection result as 91.3% in mean average precision and 19 ms per frame on GPU, compared to a RetinaNet based detection method in 86.6% and 32 ms.
Attentive CT Lesion Detection Using Deep Pyramid Inference with Multi-Scale Booster
Jul 09, 2019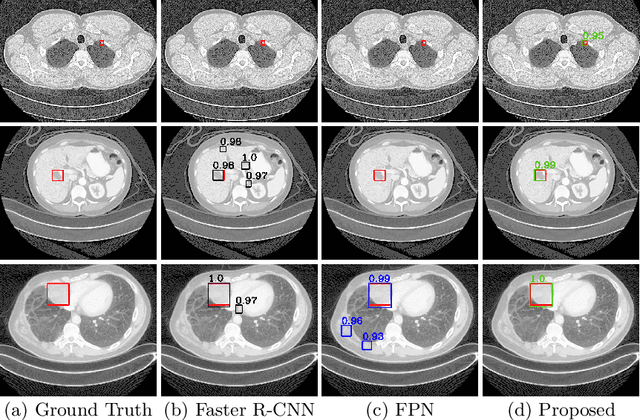
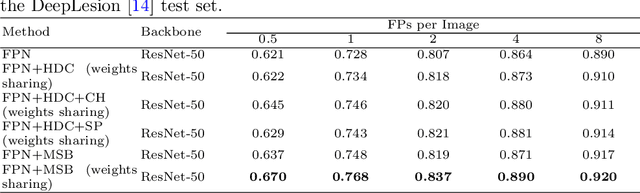
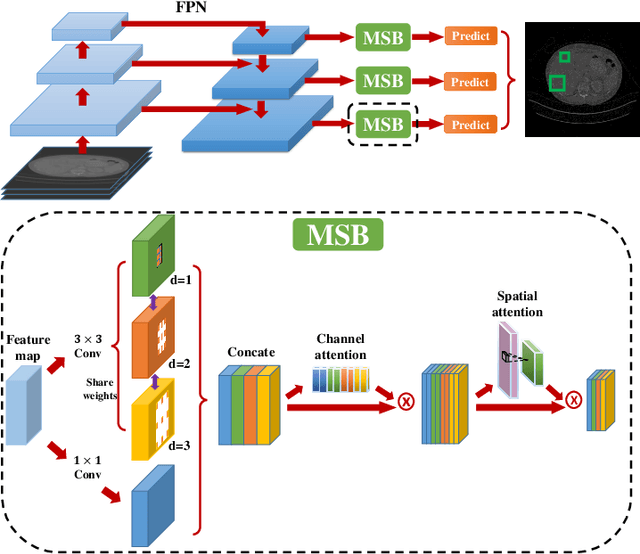
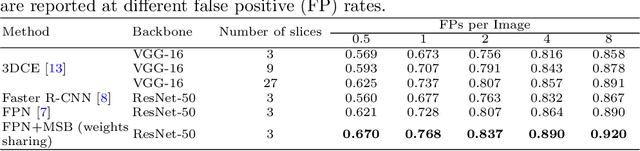
Abstract:Accurate lesion detection in computer tomography (CT) slices benefits pathologic organ analysis in the medical diagnosis process. More recently, it has been tackled as an object detection problem using the Convolutional Neural Networks (CNNs). Despite the achievements from off-the-shelf CNN models, the current detection accuracy is limited by the inability of CNNs on lesions at vastly different scales. In this paper, we propose a Multi-Scale Booster (MSB) with channel and spatial attention integrated into the backbone Feature Pyramid Network (FPN). In each pyramid level, the proposed MSB captures fine-grained scale variations by using Hierarchically Dilated Convolutions (HDC). Meanwhile, the proposed channel and spatial attention modules increase the network's capability of selecting relevant features response for lesion detection. Extensive experiments on the DeepLesion benchmark dataset demonstrate that the proposed method performs superiorly against state-of-the-art approaches.
X2CT-GAN: Reconstructing CT from Biplanar X-Rays with Generative Adversarial Networks
May 16, 2019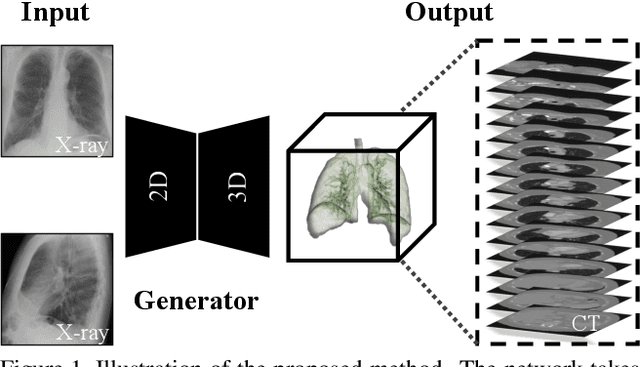
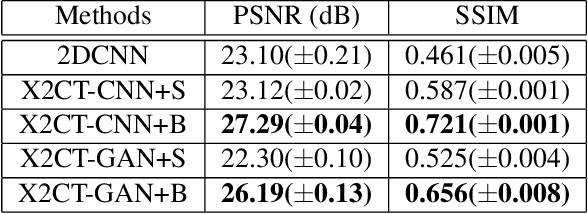
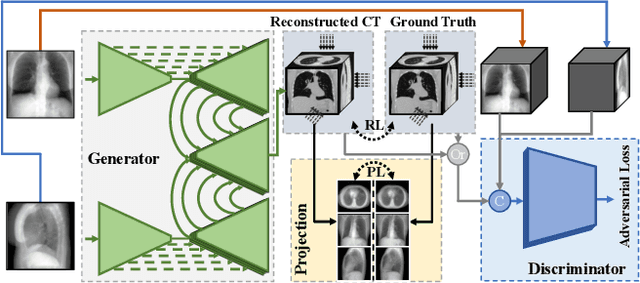
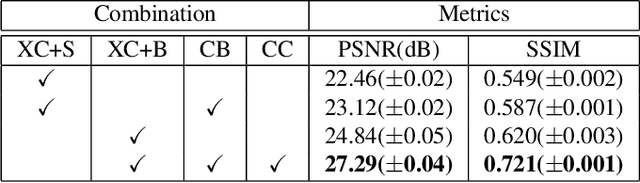
Abstract:Computed tomography (CT) can provide a 3D view of the patient's internal organs, facilitating disease diagnosis, but it incurs more radiation dose to a patient and a CT scanner is much more cost prohibitive than an X-ray machine too. Traditional CT reconstruction methods require hundreds of X-ray projections through a full rotational scan of the body, which cannot be performed on a typical X-ray machine. In this work, we propose to reconstruct CT from two orthogonal X-rays using the generative adversarial network (GAN) framework. A specially designed generator network is exploited to increase data dimension from 2D (X-rays) to 3D (CT), which is not addressed in previous research of GAN. A novel feature fusion method is proposed to combine information from two X-rays.The mean squared error (MSE) loss and adversarial loss are combined to train the generator, resulting in a high-quality CT volume both visually and quantitatively. Extensive experiments on a publicly available chest CT dataset demonstrate the effectiveness of the proposed method. It could be a nice enhancement of a low-cost X-ray machine to provide physicians a CT-like 3D volume in several niche applications.
Med3D: Transfer Learning for 3D Medical Image Analysis
Apr 09, 2019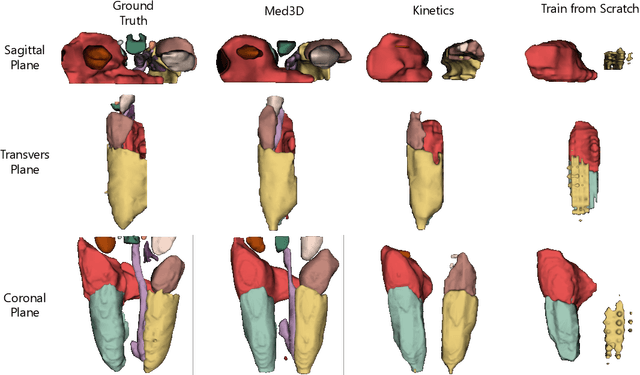
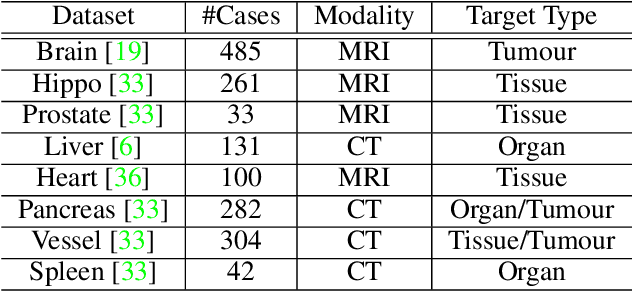
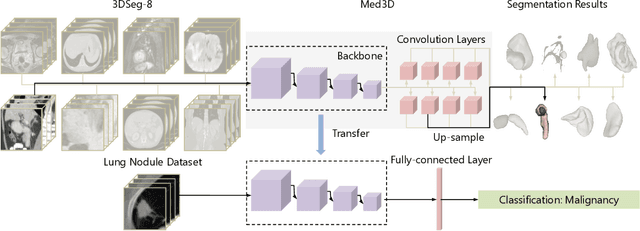
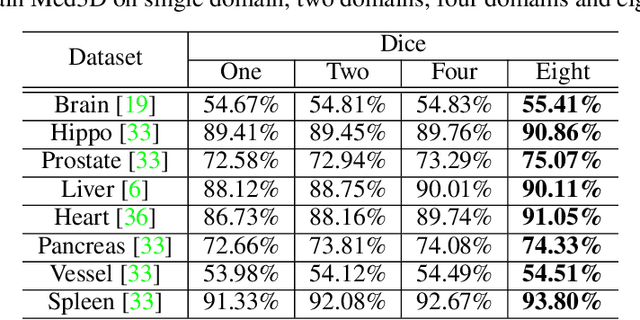
Abstract:The performance on deep learning is significantly affected by volume of training data. Models pre-trained from massive dataset such as ImageNet become a powerful weapon for speeding up training convergence and improving accuracy. Similarly, models based on large dataset are important for the development of deep learning in 3D medical images. However, it is extremely challenging to build a sufficiently large dataset due to difficulty of data acquisition and annotation in 3D medical imaging. We aggregate the dataset from several medical challenges to build 3DSeg-8 dataset with diverse modalities, target organs, and pathologies. To extract general medical three-dimension (3D) features, we design a heterogeneous 3D network called Med3D to co-train multi-domain 3DSeg-8 so as to make a series of pre-trained models. We transfer Med3D pre-trained models to lung segmentation in LIDC dataset, pulmonary nodule classification in LIDC dataset and liver segmentation on LiTS challenge. Experiments show that the Med3D can accelerate the training convergence speed of target 3D medical tasks 2 times compared with model pre-trained on Kinetics dataset, and 10 times compared with training from scratch as well as improve accuracy ranging from 3% to 20%. Transferring our Med3D model on state-the-of-art DenseASPP segmentation network, in case of single model, we achieve 94.6\% Dice coefficient which approaches the result of top-ranged algorithms on the LiTS challenge.
TAN: Temporal Affine Network for Real-Time Left Ventricle Anatomical Structure Analysis Based on 2D Ultrasound Videos
Apr 01, 2019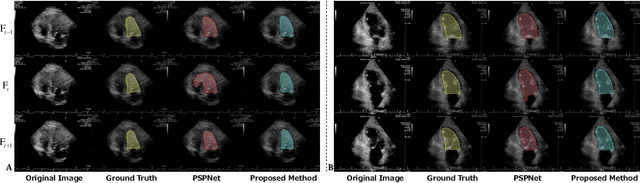
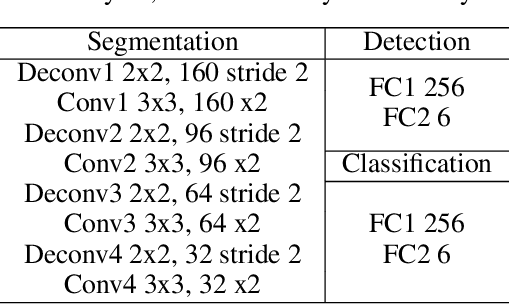
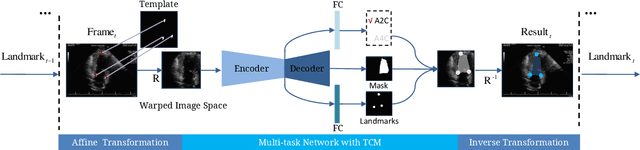
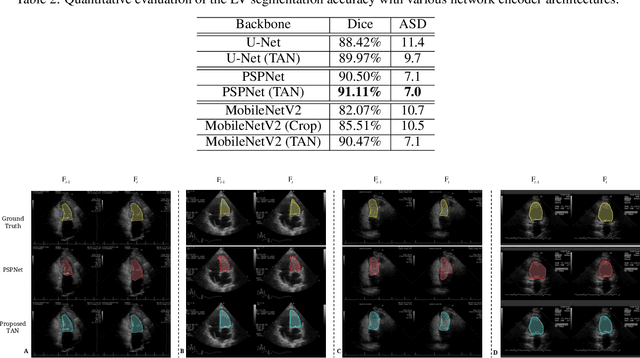
Abstract:With superiorities on low cost, portability, and free of radiation, echocardiogram is a widely used imaging modality for left ventricle (LV) function quantification. However, automatic LV segmentation and motion tracking is still a challenging task. In addition to fuzzy border definition, low contrast, and abounding artifacts on typical ultrasound images, the shape and size of the LV change significantly in a cardiac cycle. In this work, we propose a temporal affine network (TAN) to perform image analysis in a warped image space, where the shape and size variations due to the cardiac motion as well as other artifacts are largely compensated. Furthermore, we perform three frequent echocardiogram interpretation tasks simultaneously: standard cardiac plane recognition, LV landmark detection, and LV segmentation. Instead of using three networks with one dedicating to each task, we use a multi-task network to perform three tasks simultaneously. Since three tasks share the same encoder, the compact network improves the segmentation accuracy with more supervision. The network is further finetuned with optical flow adjusted annotations to enhance motion coherence in the segmentation result. Experiments on 1,714 2D echocardiographic sequences demonstrate that the proposed method achieves state-of-the-art segmentation accuracy with real-time efficiency.
Generating Synthetic X-ray Images of a Person from the Surface Geometry
May 14, 2018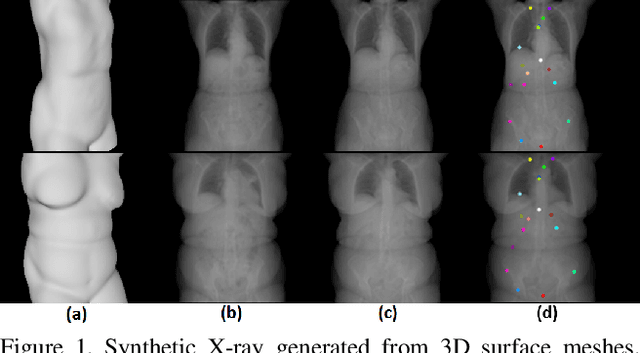
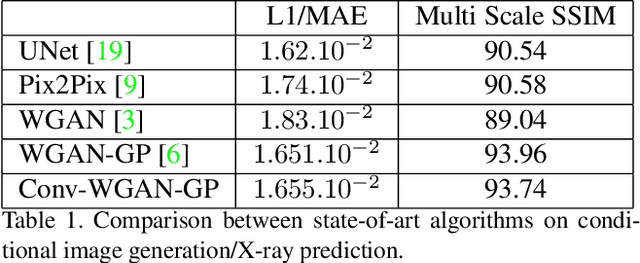
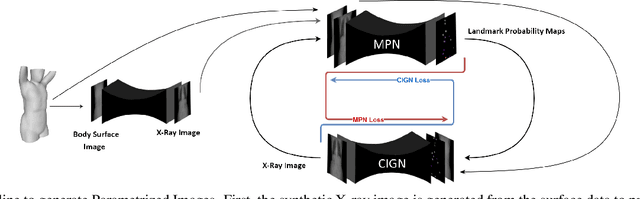
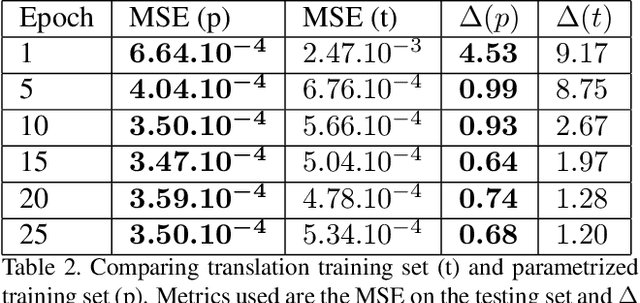
Abstract:We present a novel framework that learns to predict human anatomy from body surface. Specifically, our approach generates a synthetic X-ray image of a person only from the person's surface geometry. Furthermore, the synthetic X-ray image is parametrized and can be manipulated by adjusting a set of body markers which are also generated during the X-ray image prediction. With the proposed framework, multiple synthetic X-ray images can easily be generated by varying surface geometry. By perturbing the parameters, several additional synthetic X-ray images can be generated from the same surface geometry. As a result, our approach offers a potential to overcome the training data barrier in the medical domain. This capability is achieved by learning a pair of networks - one learns to generate the full image from the partial image and a set of parameters, and the other learns to estimate the parameters given the full image. During training, the two networks are trained iteratively such that they would converge to a solution where the predicted parameters and the full image are consistent with each other. In addition to medical data enrichment, our framework can also be used for image completion as well as anomaly detection.
DepthSynth: Real-Time Realistic Synthetic Data Generation from CAD Models for 2.5D Recognition
Nov 28, 2017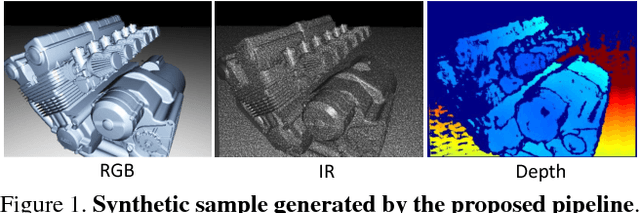
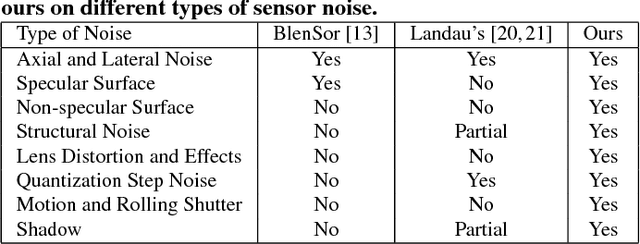
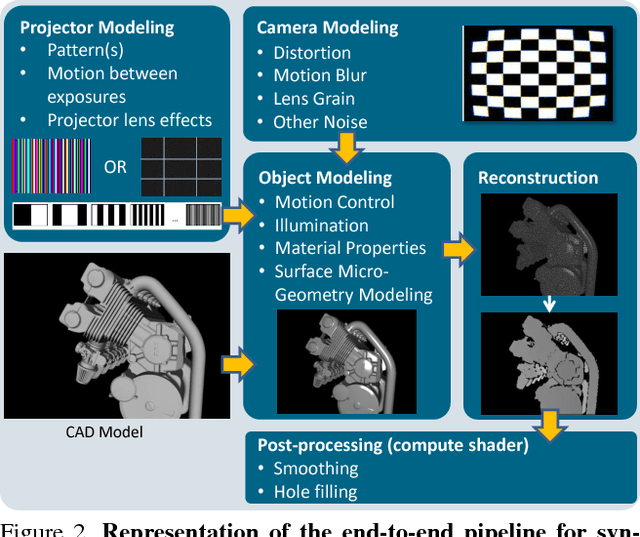
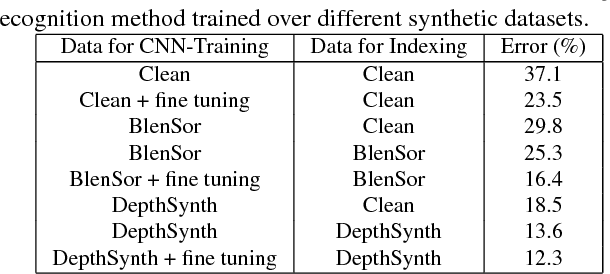
Abstract:Recent progress in computer vision has been dominated by deep neural networks trained over large amounts of labeled data. Collecting such datasets is however a tedious, often impossible task; hence a surge in approaches relying solely on synthetic data for their training. For depth images however, discrepancies with real scans still noticeably affect the end performance. We thus propose an end-to-end framework which simulates the whole mechanism of these devices, generating realistic depth data from 3D models by comprehensively modeling vital factors e.g. sensor noise, material reflectance, surface geometry. Not only does our solution cover a wider range of sensors and achieve more realistic results than previous methods, assessed through extended evaluation, but we go further by measuring the impact on the training of neural networks for various recognition tasks; demonstrating how our pipeline seamlessly integrates such architectures and consistently enhances their performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge