Justin Kirby
The Brain Tumor Segmentation Challenge 2023: Brain MR Image Synthesis for Tumor Segmentation
May 20, 2023
Abstract:Automated brain tumor segmentation methods are well established, reaching performance levels with clear clinical utility. Most algorithms require four input magnetic resonance imaging (MRI) modalities, typically T1-weighted images with and without contrast enhancement, T2-weighted images, and FLAIR images. However, some of these sequences are often missing in clinical practice, e.g., because of time constraints and/or image artifacts (such as patient motion). Therefore, substituting missing modalities to recover segmentation performance in these scenarios is highly desirable and necessary for the more widespread adoption of such algorithms in clinical routine. In this work, we report the set-up of the Brain MR Image Synthesis Benchmark (BraSyn), organized in conjunction with the Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2023. The objective of the challenge is to benchmark image synthesis methods that realistically synthesize missing MRI modalities given multiple available images to facilitate automated brain tumor segmentation pipelines. The image dataset is multi-modal and diverse, created in collaboration with various hospitals and research institutions.
The Brain Tumor Segmentation Challenge 2023: Local Synthesis of Healthy Brain Tissue via Inpainting
May 15, 2023



Abstract:A myriad of algorithms for the automatic analysis of brain MR images is available to support clinicians in their decision-making. For brain tumor patients, the image acquisition time series typically starts with a scan that is already pathological. This poses problems, as many algorithms are designed to analyze healthy brains and provide no guarantees for images featuring lesions. Examples include but are not limited to algorithms for brain anatomy parcellation, tissue segmentation, and brain extraction. To solve this dilemma, we introduce the BraTS 2023 inpainting challenge. Here, the participants' task is to explore inpainting techniques to synthesize healthy brain scans from lesioned ones. The following manuscript contains the task formulation, dataset, and submission procedure. Later it will be updated to summarize the findings of the challenge. The challenge is organized as part of the BraTS 2023 challenge hosted at the MICCAI 2023 conference in Vancouver, Canada.
Report of the Medical Image De-Identification Task Group -- Best Practices and Recommendations
Apr 01, 2023Abstract:This report addresses the technical aspects of de-identification of medical images of human subjects and biospecimens, such that re-identification risk of ethical, moral, and legal concern is sufficiently reduced to allow unrestricted public sharing for any purpose, regardless of the jurisdiction of the source and distribution sites. All medical images, regardless of the mode of acquisition, are considered, though the primary emphasis is on those with accompanying data elements, especially those encoded in formats in which the data elements are embedded, particularly Digital Imaging and Communications in Medicine (DICOM). These images include image-like objects such as Segmentations, Parametric Maps, and Radiotherapy (RT) Dose objects. The scope also includes related non-image objects, such as RT Structure Sets, Plans and Dose Volume Histograms, Structured Reports, and Presentation States. Only de-identification of publicly released data is considered, and alternative approaches to privacy preservation, such as federated learning for artificial intelligence (AI) model development, are out of scope, as are issues of privacy leakage from AI model sharing. Only technical issues of public sharing are addressed.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Lung-Originated Tumor Segmentation from Computed Tomography Scan (LOTUS) Benchmark
Jan 03, 2022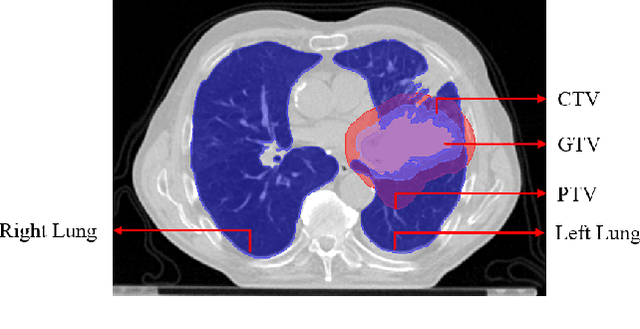
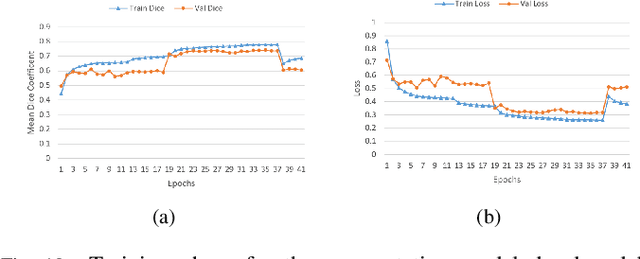
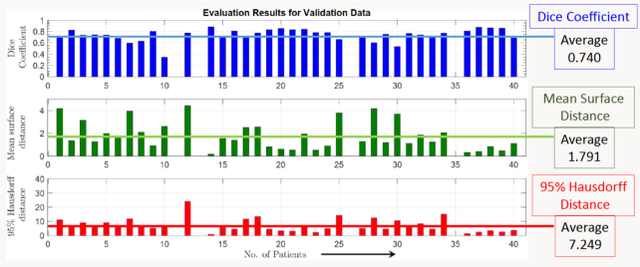
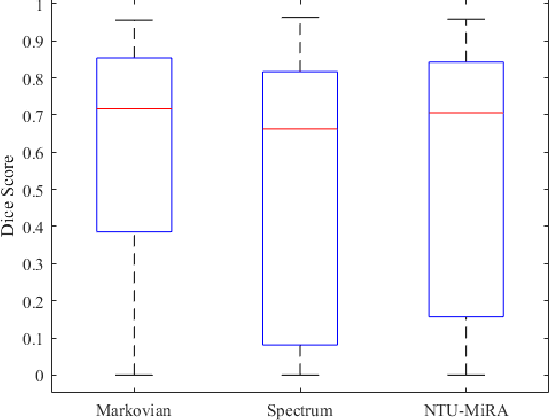
Abstract:Lung cancer is one of the deadliest cancers, and in part its effective diagnosis and treatment depend on the accurate delineation of the tumor. Human-centered segmentation, which is currently the most common approach, is subject to inter-observer variability, and is also time-consuming, considering the fact that only experts are capable of providing annotations. Automatic and semi-automatic tumor segmentation methods have recently shown promising results. However, as different researchers have validated their algorithms using various datasets and performance metrics, reliably evaluating these methods is still an open challenge. The goal of the Lung-Originated Tumor Segmentation from Computed Tomography Scan (LOTUS) Benchmark created through 2018 IEEE Video and Image Processing (VIP) Cup competition, is to provide a unique dataset and pre-defined metrics, so that different researchers can develop and evaluate their methods in a unified fashion. The 2018 VIP Cup started with a global engagement from 42 countries to access the competition data. At the registration stage, there were 129 members clustered into 28 teams from 10 countries, out of which 9 teams made it to the final stage and 6 teams successfully completed all the required tasks. In a nutshell, all the algorithms proposed during the competition, are based on deep learning models combined with a false positive reduction technique. Methods developed by the three finalists show promising results in tumor segmentation, however, more effort should be put into reducing the false positive rate. This competition manuscript presents an overview of the VIP-Cup challenge, along with the proposed algorithms and results.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021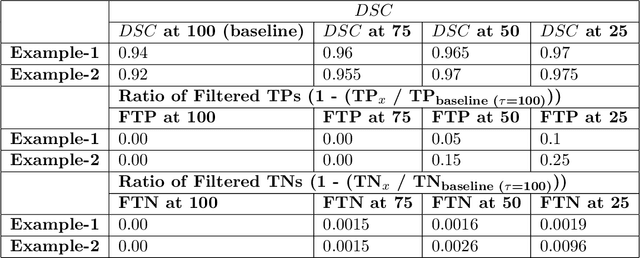
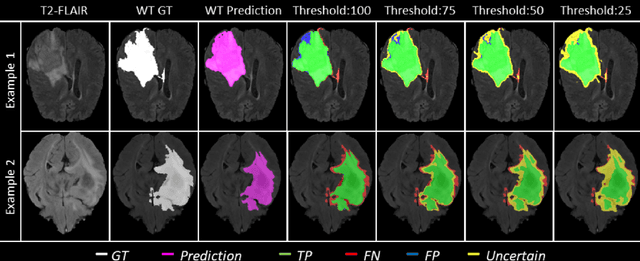
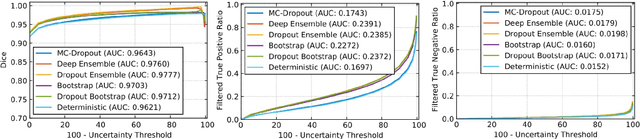
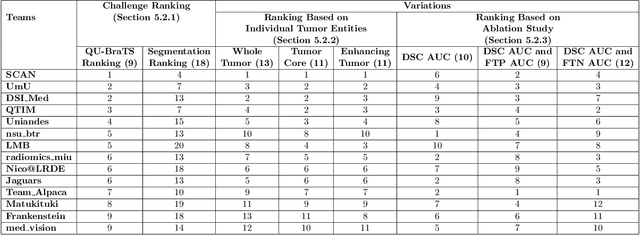
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge