Roberto Salgado
ECTIL: Label-efficient Computational Tumour Infiltrating Lymphocyte (TIL) assessment in breast cancer: Multicentre validation in 2,340 patients with breast cancer
Jan 24, 2025



Abstract:The level of tumour-infiltrating lymphocytes (TILs) is a prognostic factor for patients with (triple-negative) breast cancer (BC). Computational TIL assessment (CTA) has the potential to assist pathologists in this labour-intensive task, but current CTA models rely heavily on many detailed annotations. We propose and validate a fundamentally simpler deep learning based CTA that can be trained in only ten minutes on hundredfold fewer pathologist annotations. We collected whole slide images (WSIs) with TILs scores and clinical data of 2,340 patients with BC from six cohorts including three randomised clinical trials. Morphological features were extracted from whole slide images (WSIs) using a pathology foundation model. Our label-efficient Computational stromal TIL assessment model (ECTIL) directly regresses the TILs score from these features. ECTIL trained on only a few hundred samples (ECTIL-TCGA) showed concordance with the pathologist over five heterogeneous external cohorts (r=0.54-0.74, AUROC=0.80-0.94). Training on all slides of five cohorts (ECTIL-combined) improved results on a held-out test set (r=0.69, AUROC=0.85). Multivariable Cox regression analyses indicated that every 10% increase of ECTIL scores was associated with improved overall survival independent of clinicopathological variables (HR 0.86, p<0.01), similar to the pathologist score (HR 0.87, p<0.001). We demonstrate that ECTIL is highly concordant with an expert pathologist and obtains a similar hazard ratio. ECTIL has a fundamentally simpler design than existing methods and can be trained on orders of magnitude fewer annotations. Such a CTA may be used to pre-screen patients for, e.g., immunotherapy clinical trial inclusion, or as a tool to assist clinicians in the diagnostic work-up of patients with BC. Our model is available under an open source licence (https://github.com/nki-ai/ectil).
Automated Scoring of Nuclear Pleomorphism Spectrum with Pathologist-level Performance in Breast Cancer
Dec 24, 2020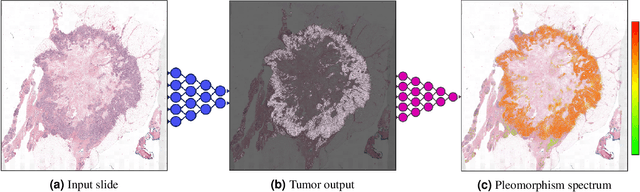
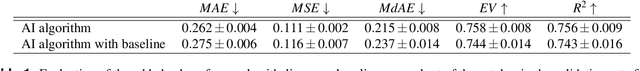
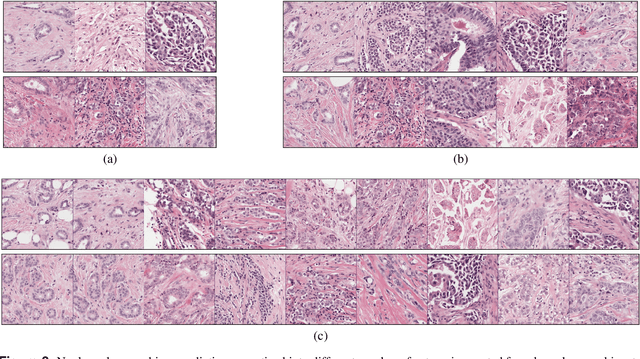
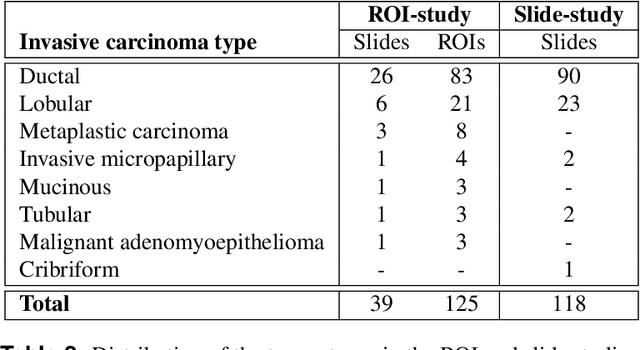
Abstract:Nuclear pleomorphism, defined herein as the extent of abnormalities in the overall appearance of tumor nuclei, is one of the components of the three-tiered breast cancer grading. Given that nuclear pleomorphism reflects a continuous spectrum of variation, we trained a deep neural network on a large variety of tumor regions from the collective knowledge of several pathologists, without constraining the network to the traditional three-category classification. We also motivate an additional approach in which we discuss the additional benefit of normal epithelium as baseline, following the routine clinical practice where pathologists are trained to score nuclear pleomorphism in tumor, having the normal breast epithelium for comparison. In multiple experiments, our fully-automated approach could achieve top pathologist-level performance in select regions of interest as well as at whole slide images, compared to ten and four pathologists, respectively.
Self-Supervised Nuclei Segmentation in Histopathological Images Using Attention
Jul 16, 2020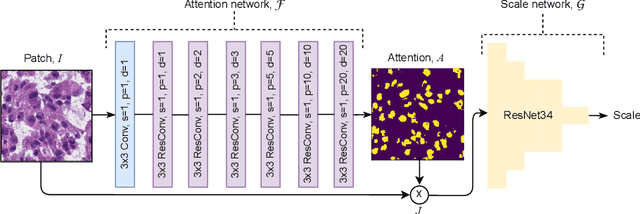
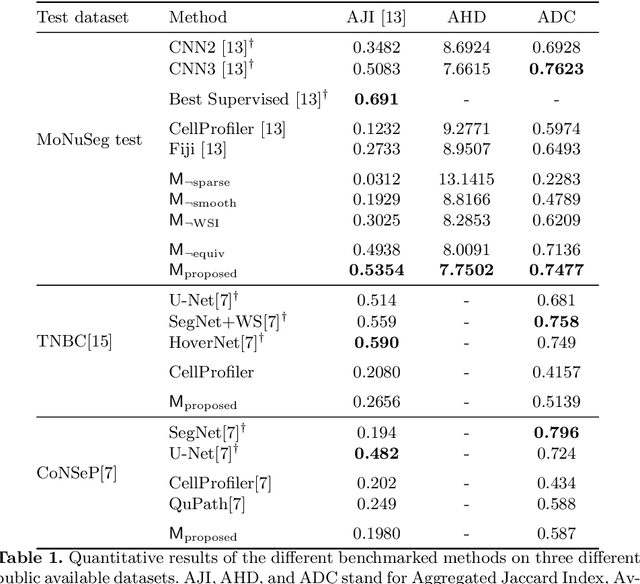
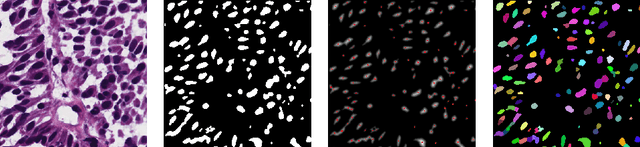
Abstract:Segmentation and accurate localization of nuclei in histopathological images is a very challenging problem, with most existing approaches adopting a supervised strategy. These methods usually rely on manual annotations that require a lot of time and effort from medical experts. In this study, we present a self-supervised approach for segmentation of nuclei for whole slide histopathology images. Our method works on the assumption that the size and texture of nuclei can determine the magnification at which a patch is extracted. We show that the identification of the magnification level for tiles can generate a preliminary self-supervision signal to locate nuclei. We further show that by appropriately constraining our model it is possible to retrieve meaningful segmentation maps as an auxiliary output to the primary magnification identification task. Our experiments show that with standard post-processing, our method can outperform other unsupervised nuclei segmentation approaches and report similar performance with supervised ones on the publicly available MoNuSeg dataset. Our code and models are available online to facilitate further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge