Eun Sook Ko
RadiomicsFill-Mammo: Synthetic Mammogram Mass Manipulation with Radiomics Features
Jul 08, 2024Abstract:Motivated by the question, "Can we generate tumors with desired attributes?'' this study leverages radiomics features to explore the feasibility of generating synthetic tumor images. Characterized by its low-dimensional yet biologically meaningful markers, radiomics bridges the gap between complex medical imaging data and actionable clinical insights. We present RadiomicsFill-Mammo, the first of the RadiomicsFill series, an innovative technique that generates realistic mammogram mass images mirroring specific radiomics attributes using masked images and opposite breast images, leveraging a recent stable diffusion model. This approach also allows for the incorporation of essential clinical variables, such as BI-RADS and breast density, alongside radiomics features as conditions for mass generation. Results indicate that RadiomicsFill-Mammo effectively generates diverse and realistic tumor images based on various radiomics conditions. Results also demonstrate a significant improvement in mass detection capabilities, leveraging RadiomicsFill-Mammo as a strategy to generate simulated samples. Furthermore, RadiomicsFill-Mammo not only advances medical imaging research but also opens new avenues for enhancing treatment planning and tumor simulation. Our code is available at https://github.com/nainye/RadiomicsFill.
Deep learning achieves radiologist-level performance of tumor segmentation in breast MRI
Sep 21, 2020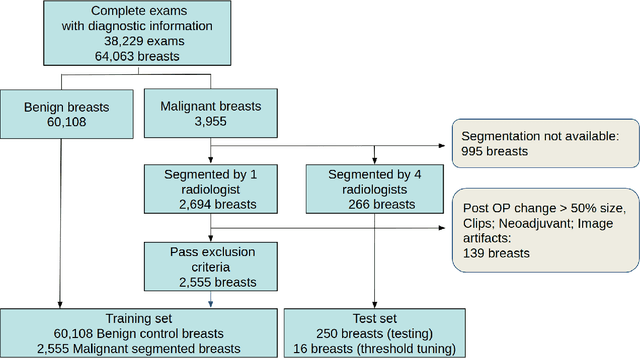
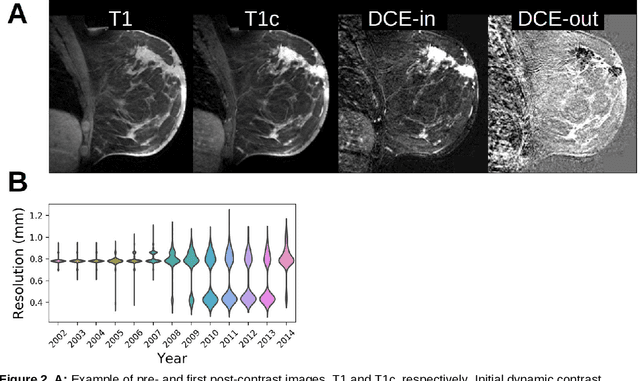
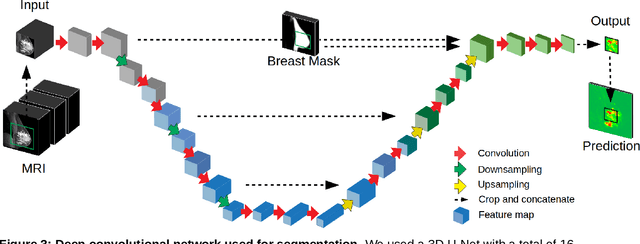
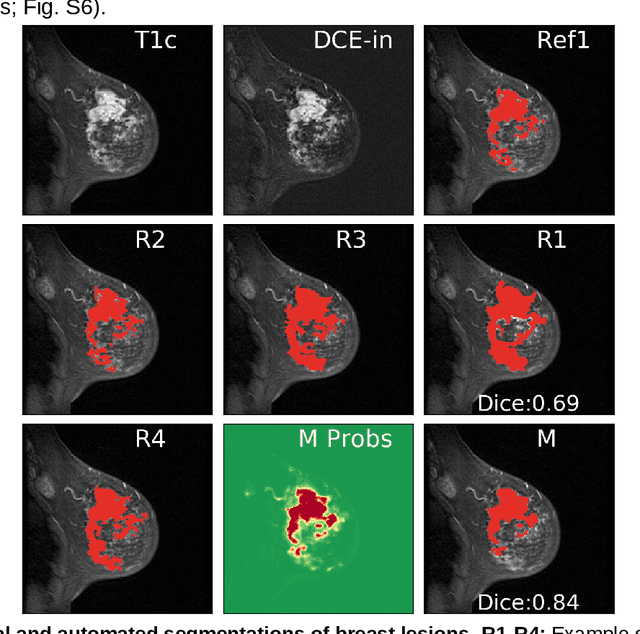
Abstract:Purpose: The goal of this research was to develop a deep network architecture that achieves fully-automated radiologist-level segmentation of breast tumors in MRI. Materials and Methods: We leveraged 38,229 clinical MRI breast exams collected retrospectively from women aged 12-94 (mean age 54) who presented between 2002 and 2014 at a single clinical site. The training set for the network consisted of 2,555 malignant breasts that were segmented in 2D by experienced radiologists, as well as 60,108 benign breasts that served as negative controls. The test set consisted of 250 exams with tumors segmented independently by four radiologists. We selected among several 3D deep convolutional neural network architectures, input modalities and harmonization methods. The outcome measure was the Dice score for 2D segmentation, and was compared between the network and radiologists using the Wilcoxon signed-rank test and the TOST procedure. Results: The best-performing network on the training set was a volumetric U-Net with contrast enhancement dynamic as input and with intensity normalized for each exam. In the test set the median Dice score of this network was 0.77. The performance of the network was equivalent to that of the radiologists (TOST procedure with radiologist performance of 0.69-0.84 as equivalence bounds: p = 5e-10 and p = 2e-5, respectively; N = 250) and compares favorably with published state of the art (0.6-0.77). Conclusion: When trained on a dataset of over 60 thousand breasts, a volumetric U-Net performs as well as expert radiologists at segmenting malignant breast lesions in MRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge