Holger Roth
3D Semi-Supervised Learning with Uncertainty-Aware Multi-View Co-Training
Nov 29, 2018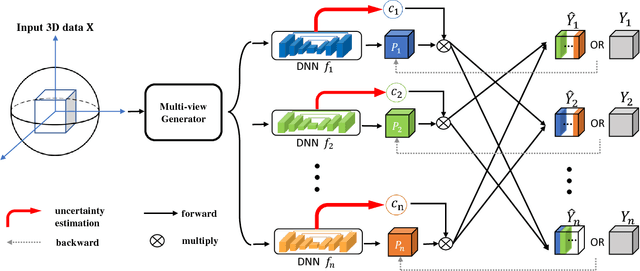
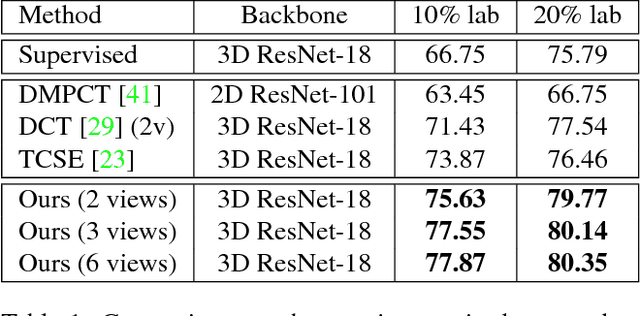
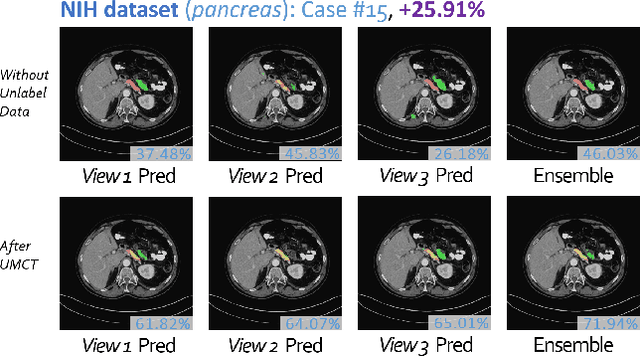
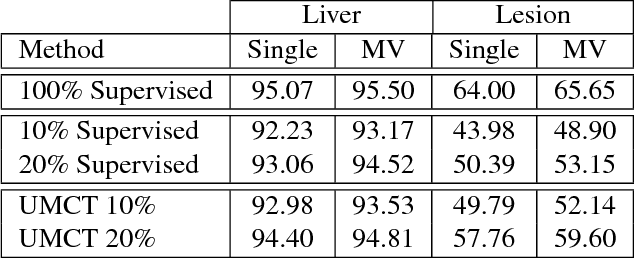
Abstract:We propose a novel framework, uncertainty-aware multi-view co-training (UMCT), to address semi-supervised learning on 3D data, such as volumetric data in medical imaging. The original co-training method was applied to non-visual data. It requires different sources, or representations, of the data, which are called different views and differ from viewpoint in computer vision. Co-training was recently applied to visual tasks where the views were deep networks learnt by adversarial training. In our work, targeted at 3D data, co-training is achieved by exploiting multi-viewpoint consistency. We generate different views by rotating the 3D data and utilize asymmetrical 3D kernels to further encourage diversified features of each sub-net. In addition, we propose an uncertainty-aware attention mechanism to estimate the reliability of each view prediction with Bayesian deep learning. As one view requires the supervision from other views in co-training, our self-adaptive approach computes a confidence score for the prediction of each unlabeled sample, in order to assign a reliable pseudo label and thus achieve better performance. We show the effectiveness of our proposed method on several open datasets from medical image segmentation tasks (NIH pancreas & LiTS liver tumor dataset). A method based on our approach achieved the state-of-the-art performances on both the LiTS liver tumor segmentation and the Medical Segmentation Decathlon (MSD) challenge, demonstrating the robustness and value of our framework even when fully supervised training is feasible.
Towards dense volumetric pancreas segmentation in CT using 3D fully convolutional networks
Jan 19, 2018Abstract:Pancreas segmentation in computed tomography imaging has been historically difficult for automated methods because of the large shape and size variations between patients. In this work, we describe a custom-build 3D fully convolutional network (FCN) that can process a 3D image including the whole pancreas and produce an automatic segmentation. We investigate two variations of the 3D FCN architecture; one with concatenation and one with summation skip connections to the decoder part of the network. We evaluate our methods on a dataset from a clinical trial with gastric cancer patients, including 147 contrast enhanced abdominal CT scans acquired in the portal venous phase. Using the summation architecture, we achieve an average Dice score of 89.7 $\pm$ 3.8 (range [79.8, 94.8]) % in testing, achieving the new state-of-the-art performance in pancreas segmentation on this dataset.
* Accepted for oral presentation at SPIE Medical Imaging 2018, Houston, TX, USA Updated experiment in Fig. 4
Towards Automatic Abdominal Multi-Organ Segmentation in Dual Energy CT using Cascaded 3D Fully Convolutional Network
Oct 15, 2017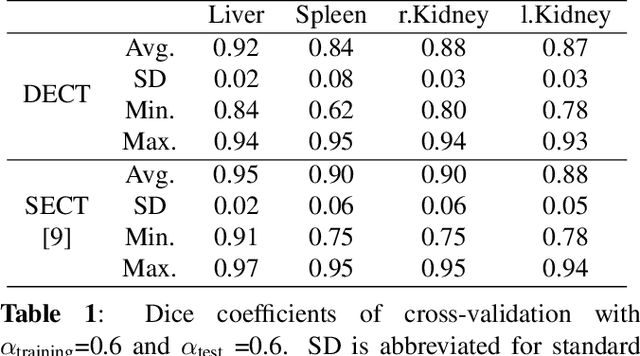
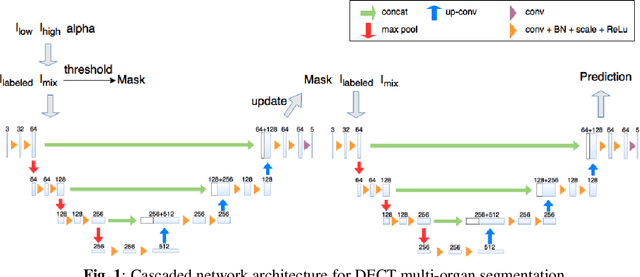
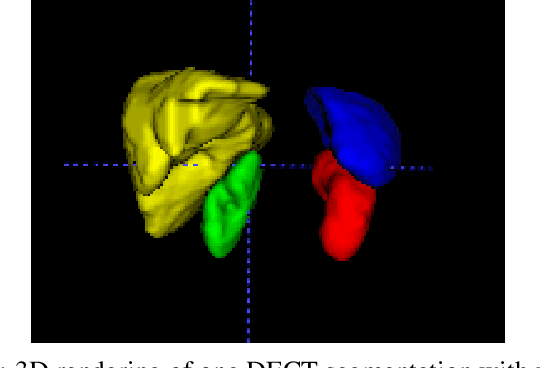
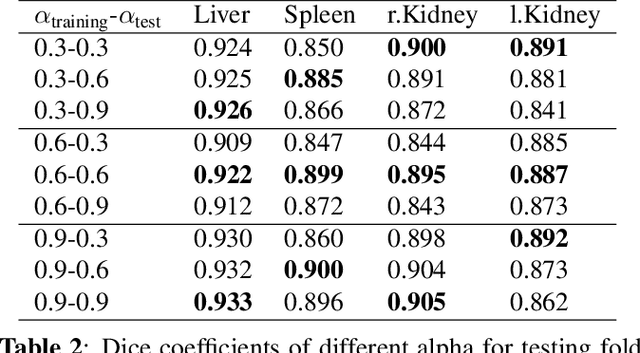
Abstract:Automatic multi-organ segmentation of the dual energy computed tomography (DECT) data can be beneficial for biomedical research and clinical applications. However, it is a challenging task. Recent advances in deep learning showed the feasibility to use 3-D fully convolutional networks (FCN) for voxel-wise dense predictions in single energy computed tomography (SECT). In this paper, we proposed a 3D FCN based method for automatic multi-organ segmentation in DECT. The work was based on a cascaded FCN and a general model for the major organs trained on a large set of SECT data. We preprocessed the DECT data by using linear weighting and fine-tuned the model for the DECT data. The method was evaluated using 42 torso DECT data acquired with a clinical dual-source CT system. Four abdominal organs (liver, spleen, left and right kidneys) were evaluated. Cross-validation was tested. Effect of the weight on the accuracy was researched. In all the tests, we achieved an average Dice coefficient of 93% for the liver, 90% for the spleen, 91% for the right kidney and 89% for the left kidney, respectively. The results show our method is feasible and promising.
2D View Aggregation for Lymph Node Detection Using a Shallow Hierarchy of Linear Classifiers
Aug 14, 2014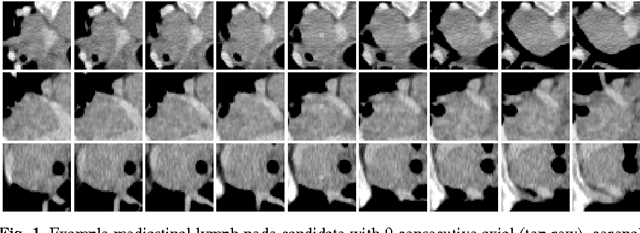

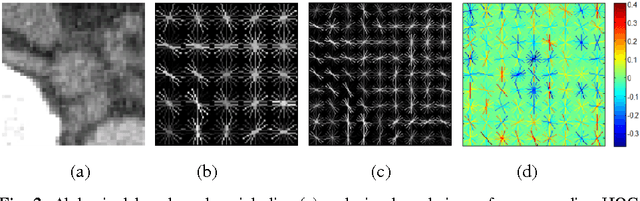
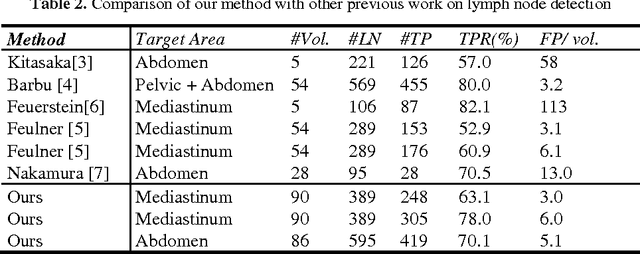
Abstract:Enlarged lymph nodes (LNs) can provide important information for cancer diagnosis, staging, and measuring treatment reactions, making automated detection a highly sought goal. In this paper, we propose a new algorithm representation of decomposing the LN detection problem into a set of 2D object detection subtasks on sampled CT slices, largely alleviating the curse of dimensionality issue. Our 2D detection can be effectively formulated as linear classification on a single image feature type of Histogram of Oriented Gradients (HOG), covering a moderate field-of-view of 45 by 45 voxels. We exploit both simple pooling and sparse linear fusion schemes to aggregate these 2D detection scores for the final 3D LN detection. In this manner, detection is more tractable and does not need to perform perfectly at instance level (as weak hypotheses) since our aggregation process will robustly harness collective information for LN detection. Two datasets (90 patients with 389 mediastinal LNs and 86 patients with 595 abdominal LNs) are used for validation. Cross-validation demonstrates 78.0% sensitivity at 6 false positives/volume (FP/vol.) (86.1% at 10 FP/vol.) and 73.1% sensitivity at 6 FP/vol. (87.2% at 10 FP/vol.), for the mediastinal and abdominal datasets respectively. Our results compare favorably to previous state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge