Fuping Wu
Decoupling Predictions in Distributed Learning for Multi-Center Left Atrial MRI Segmentation
Jun 10, 2022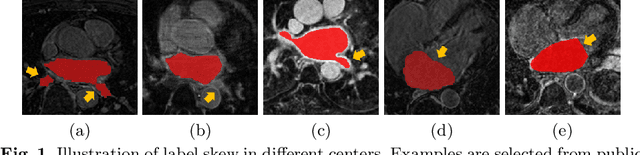
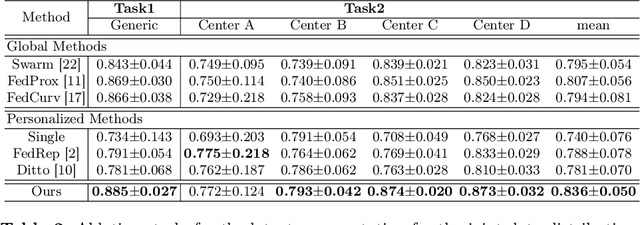
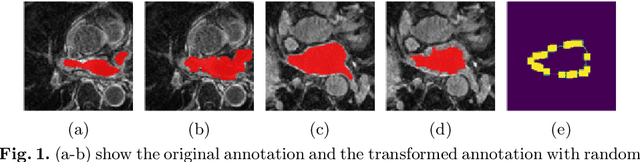
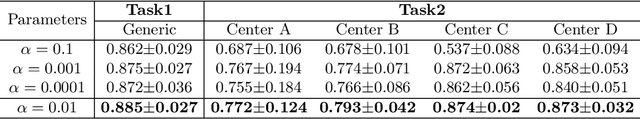
Abstract:Distributed learning has shown great potential in medical image analysis. It allows to use multi-center training data with privacy protection. However, data distributions in local centers can vary from each other due to different imaging vendors, and annotation protocols. Such variation degrades the performance of learning-based methods. To mitigate the influence, two groups of methods have been proposed for different aims, i.e., the global methods and the personalized methods. The former are aimed to improve the performance of a single global model for all test data from unseen centers (known as generic data); while the latter target multiple models for each center (denoted as local data). However, little has been researched to achieve both goals simultaneously. In this work, we propose a new framework of distributed learning that bridges the gap between two groups, and improves the performance for both generic and local data. Specifically, our method decouples the predictions for generic data and local data, via distribution-conditioned adaptation matrices. Results on multi-center left atrial (LA) MRI segmentation showed that our method demonstrated superior performance over existing methods on both generic and local data. Our code is available at https://github.com/key1589745/decouple_predict
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022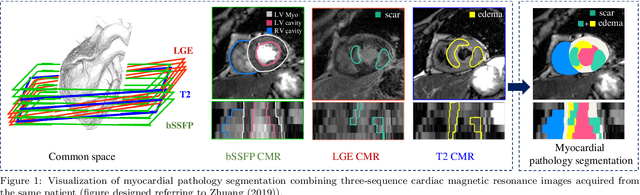
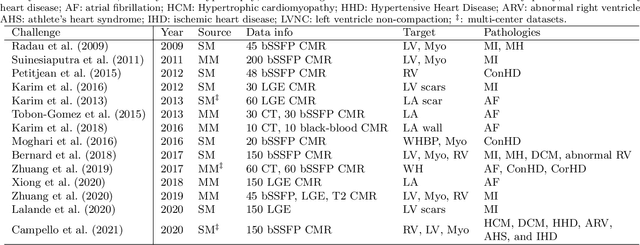
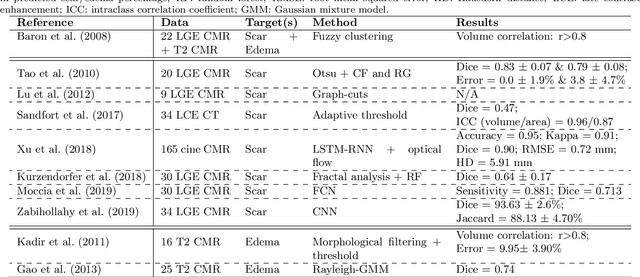
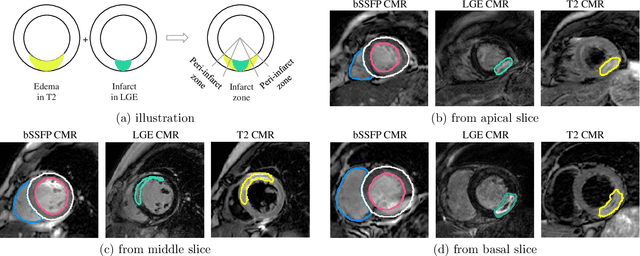
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
Multi-Modality Cardiac Image Analysis with Deep Learning
Nov 08, 2021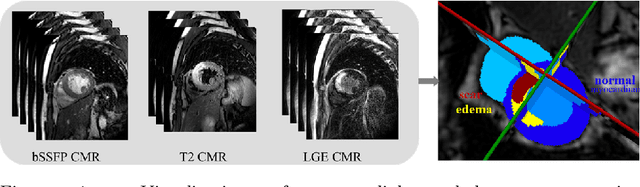

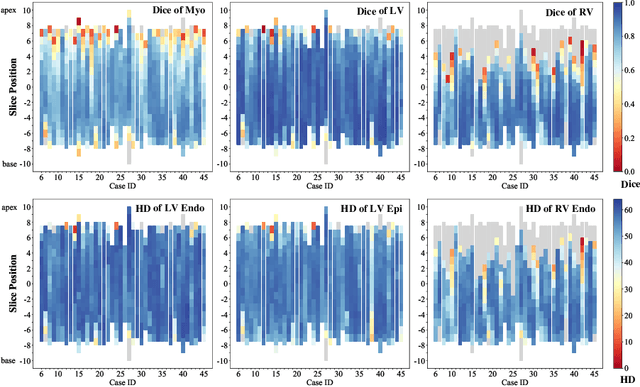
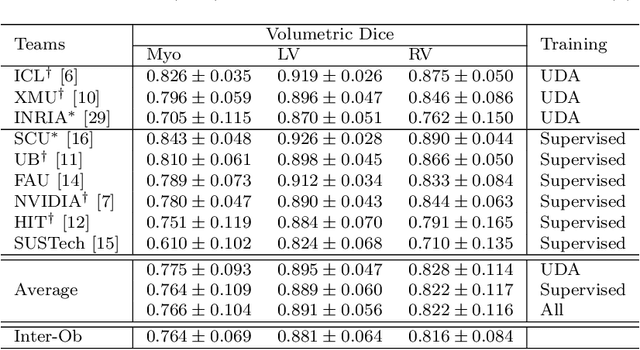
Abstract:Accurate cardiac computing, analysis and modeling from multi-modality images are important for the diagnosis and treatment of cardiac disease. Late gadolinium enhancement magnetic resonance imaging (LGE MRI) is a promising technique to visualize and quantify myocardial infarction (MI) and atrial scars. Automating quantification of MI and atrial scars can be challenging due to the low image quality and complex enhancement patterns of LGE MRI. Moreover, compared with the other sequences LGE MRIs with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation and quantification of LGE MRIs. This chapter aims to summarize the state-of-the-art and our recent advanced contributions on deep learning based multi-modality cardiac image analysis. Firstly, we introduce two benchmark works for multi-sequence cardiac MRI based myocardial and pathology segmentation. Secondly, two novel frameworks for left atrial scar segmentation and quantification from LGE MRI were presented. Thirdly, we present three unsupervised domain adaptation techniques for cross-modality cardiac image segmentation.
Unsupervised Domain Adaptation with Variational Approximation for Cardiac Segmentation
Jun 16, 2021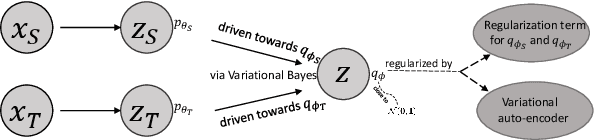
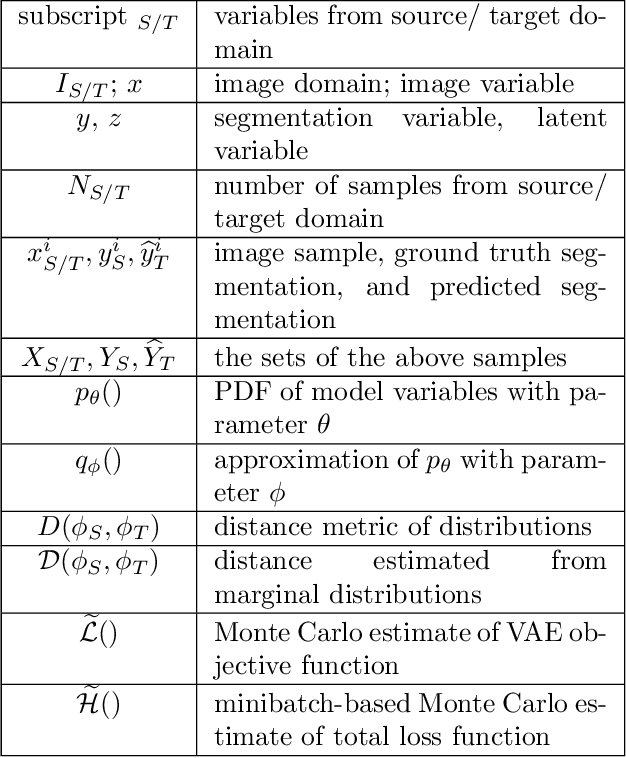
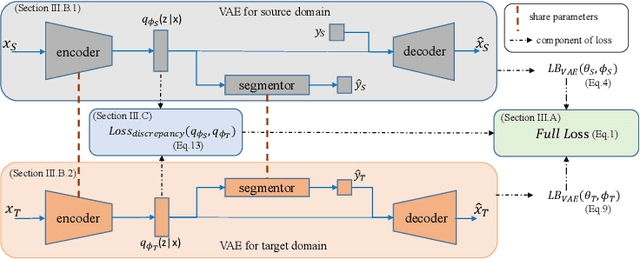
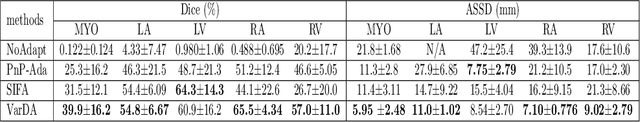
Abstract:Unsupervised domain adaptation is useful in medical image segmentation. Particularly, when ground truths of the target images are not available, domain adaptation can train a target-specific model by utilizing the existing labeled images from other modalities. Most of the reported works mapped images of both the source and target domains into a common latent feature space, and then reduced their discrepancy either implicitly with adversarial training or explicitly by directly minimizing a discrepancy metric. In this work, we propose a new framework, where the latent features of both domains are driven towards a common and parameterized variational form, whose conditional distribution given the image is Gaussian. This is achieved by two networks based on variational auto-encoders (VAEs) and a regularization for this variational approximation. Both of the VAEs, each for one domain, contain a segmentation module, where the source segmentation is trained in a supervised manner, while the target one is trained unsupervisedly. We validated the proposed domain adaptation method using two cardiac segmentation tasks, i.e., the cross-modality (CT and MR) whole heart segmentation and the cross-sequence cardiac MR segmentation. Results show that the proposed method achieved better accuracies compared to two state-of-the-art approaches and demonstrated good potential for cardiac segmentation. Furthermore, the proposed explicit regularization was shown to be effective and efficient in narrowing down the distribution gap between domains, which is useful for unsupervised domain adaptation. Our code and data has been released via https://zmiclab.github.io/projects.html.
Random Style Transfer based Domain Generalization Networks Integrating Shape and Spatial Information
Sep 03, 2020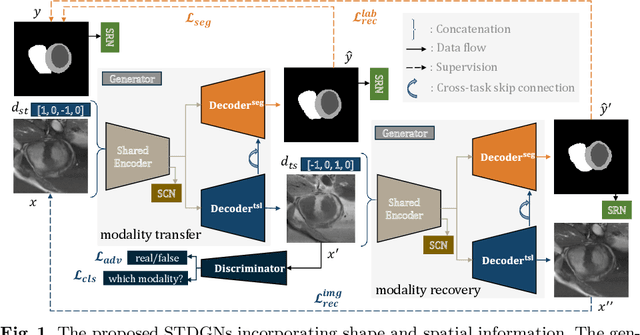
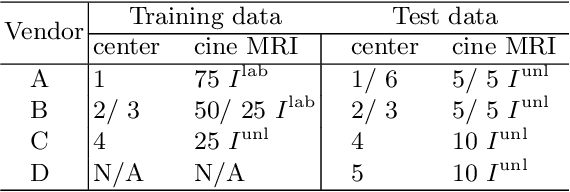
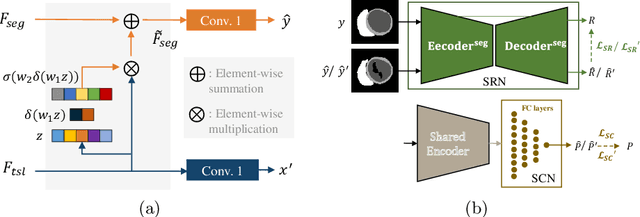
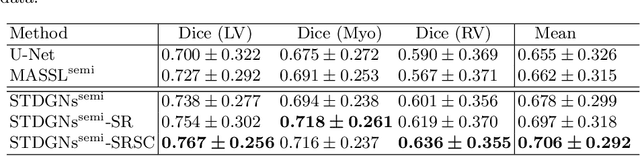
Abstract:Deep learning (DL)-based models have demonstrated good performance in medical image segmentation. However, the models trained on a known dataset often fail when performed on an unseen dataset collected from different centers, vendors and disease populations. In this work, we present a random style transfer network to tackle the domain generalization problem for multi-vendor and center cardiac image segmentation. Style transfer is used to generate training data with a wider distribution/ heterogeneity, namely domain augmentation. As the target domain could be unknown, we randomly generate a modality vector for the target modality in the style transfer stage, to simulate the domain shift for unknown domains. The model can be trained in a semi-supervised manner by simultaneously optimizing a supervised segmentation and an unsupervised style translation objective. Besides, the framework incorporates the spatial information and shape prior of the target by introducing two regularization terms. We evaluated the proposed framework on 40 subjects from the M\&Ms challenge2020, and obtained promising performance in the segmentation for data from unknown vendors and centers.
Atrial Scar Quantification via Multi-scale CNN in the Graph-cuts Framework
Feb 21, 2019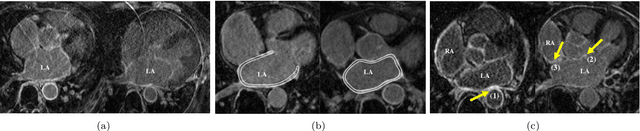
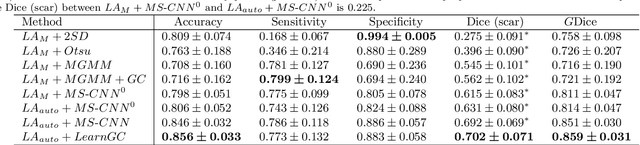
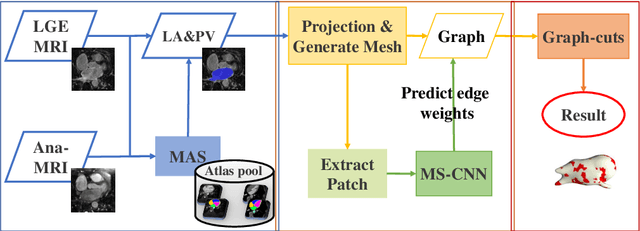

Abstract:Late gadolinium enhancement magnetic resonance imaging (LGE MRI) appears to be a promising alternative for scar assessment in patients with atrial fibrillation (AF). Automating the quantification and analysis of atrial scars can be challenging due to the low image quality. In this work, we propose a fully automated method based on the graph-cuts framework, where the potentials of the graph are learned on a surface mesh of the left atrium (LA) using a multi-scale convolutional neural network (MS-CNN). For validation, we have employed fifty-eight images with manual delineations. MS-CNN, which can efficiently incorporate both the local and global texture information of the images, has been shown to evidently improve the segmentation accuracy of the proposed graph-cuts based method. The segmentation could be further improved when the contribution between the t-link and n-link weights of the graph is balanced. The proposed method achieves a mean accuracy of 0.856 +- 0.033 and mean Dice score of 0.702 +- 0.071 for LA scar quantification. Compared with the conventional methods, which are based on the manual delineation of LA for initialization, our method is fully automatic and has demonstrated significantly better Dice score and accuracy (p < 0.01). The method is promising and can be useful in diagnosis and prognosis of AF.
Atrial scars segmentation via potential learning in the graph-cuts framework
Oct 22, 2018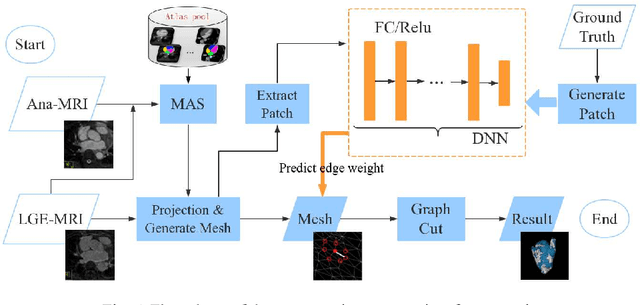
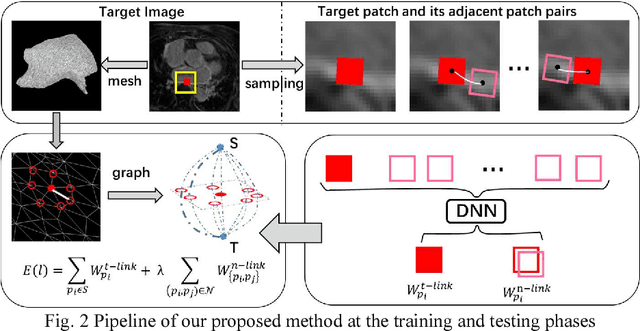
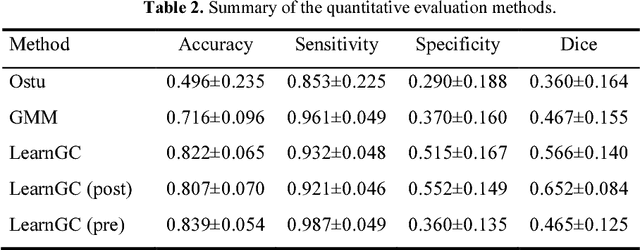
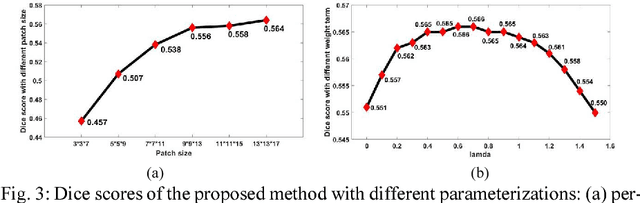
Abstract:Late Gadolinium Enhancement Magnetic Resonance Imaging (LGE MRI) emerged as a routine scan for patients with atrial fibrillation (AF). However, due to the low image quality automating the quantification and analysis of the atrial scars is challenging. In this study, we pro-posed a fully automated method based on the graph-cuts framework, where the potential of the graph is learned on a surface mesh of the left atrium (LA) using an equidistant projection and a Deep Neural Network (DNN). For validation, we employed 100 datasets with manual delineation. The results showed that the performance of the proposed method improved and converged with respect to the increased size of training patches, which provide important features of the structural and texture information learned by the DNN. The segmentation could be further improved when the contribution from the t-link and n-link is balanced, thanks to inter-relationship learned by the DNN for the graph-cuts algorithm. Compared with the published methods which mostly acquired manual delineation of the LA or LA wall, our method is fully automatic and demonstrated evidently better results with statistical significance. Finally, the accuracy of quantifying the scars assessed by the Dice score was 0.570. The results are promising and the method can be useful in diagnosis and prognosis of AF.
Atrial fibrosis quantification based on maximum likelihood estimator of multivariate images
Oct 22, 2018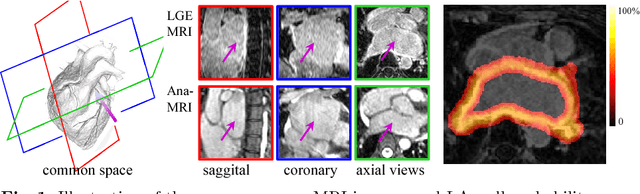
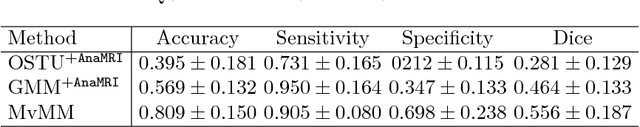
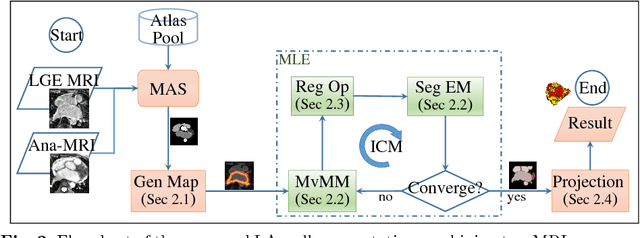
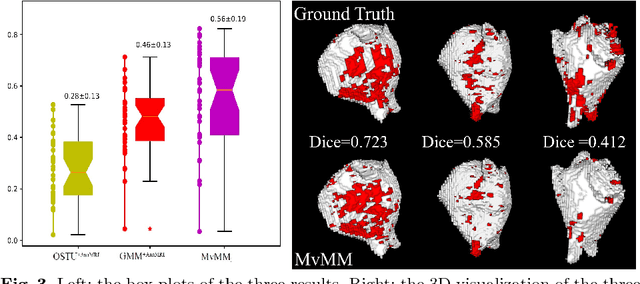
Abstract:We present a fully-automated segmentation and quantification of the left atrial (LA) fibrosis and scars combining two cardiac MRIs, one is the target late gadolinium-enhanced (LGE) image, and the other is an anatomical MRI from the same acquisition session. We formulate the joint distribution of images using a multivariate mixture model (MvMM), and employ the maximum likelihood estimator (MLE) for texture classification of the images simultaneously. The MvMM can also embed transformations assigned to the images to correct the misregistration. The iterated conditional mode algorithm is adopted for optimization. This method first extracts the anatomical shape of the LA, and then estimates a prior probability map. It projects the resulting segmentation onto the LA surface, for quantification and analysis of scarring. We applied the proposed method to 36 clinical data sets and obtained promising results (Accuracy: $0.809\pm .150$, Dice: $0.556\pm.187$). We compared the method with the conventional algorithms and showed an evidently and statistically better performance ($p<0.03$).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge