Dosik Hwang
HP-GAN: Harnessing pretrained networks for GAN improvement with FakeTwins and discriminator consistency
Feb 03, 2026Abstract:Generative Adversarial Networks (GANs) have made significant progress in enhancing the quality of image synthesis. Recent methods frequently leverage pretrained networks to calculate perceptual losses or utilize pretrained feature spaces. In this paper, we extend the capabilities of pretrained networks by incorporating innovative self-supervised learning techniques and enforcing consistency between discriminators during GAN training. Our proposed method, named HP-GAN, effectively exploits neural network priors through two primary strategies: FakeTwins and discriminator consistency. FakeTwins leverages pretrained networks as encoders to compute a self-supervised loss and applies this through the generated images to train the generator, thereby enabling the generation of more diverse and high quality images. Additionally, we introduce a consistency mechanism between discriminators that evaluate feature maps extracted from Convolutional Neural Network (CNN) and Vision Transformer (ViT) feature networks. Discriminator consistency promotes coherent learning among discriminators and enhances training robustness by aligning their assessments of image quality. Our extensive evaluation across seventeen datasets-including scenarios with large, small, and limited data, and covering a variety of image domains-demonstrates that HP-GAN consistently outperforms current state-of-the-art methods in terms of Fréchet Inception Distance (FID), achieving significant improvements in image diversity and quality. Code is available at: https://github.com/higun2/HP-GAN.
Large Language Models are Clinical Reasoners: Reasoning-Aware Diagnosis Framework with Prompt-Generated Rationales
Dec 12, 2023Abstract:Machine reasoning has made great progress in recent years owing to large language models (LLMs). In the clinical domain, however, most NLP-driven projects mainly focus on clinical classification or reading comprehension, and under-explore clinical reasoning for disease diagnosis due to the expensive rationale annotation with clinicians. In this work, we present a ``reasoning-aware'' diagnosis framework that rationalizes the diagnostic process via prompt-based learning in a time- and labor-efficient manner, and learns to reason over the prompt-generated rationales. Specifically, we address the clinical reasoning for disease diagnosis, where the LLM generates diagnostic rationales providing its insight on presented patient data and the reasoning path towards the diagnosis, namely Clinical Chain-of-Thought (Clinical CoT). We empirically demonstrate LLMs/LMs' ability of clinical reasoning via extensive experiments and analyses on both rationale generation and disease diagnosis in various settings. We further propose a novel set of criteria for evaluating machine-generated rationales' potential for real-world clinical settings, facilitating and benefiting future research in this area.
SDC-UDA: Volumetric Unsupervised Domain Adaptation Framework for Slice-Direction Continuous Cross-Modality Medical Image Segmentation
May 18, 2023Abstract:Recent advances in deep learning-based medical image segmentation studies achieve nearly human-level performance in fully supervised manner. However, acquiring pixel-level expert annotations is extremely expensive and laborious in medical imaging fields. Unsupervised domain adaptation (UDA) can alleviate this problem, which makes it possible to use annotated data in one imaging modality to train a network that can successfully perform segmentation on target imaging modality with no labels. In this work, we propose SDC-UDA, a simple yet effective volumetric UDA framework for slice-direction continuous cross-modality medical image segmentation which combines intra- and inter-slice self-attentive image translation, uncertainty-constrained pseudo-label refinement, and volumetric self-training. Our method is distinguished from previous methods on UDA for medical image segmentation in that it can obtain continuous segmentation in the slice direction, thereby ensuring higher accuracy and potential in clinical practice. We validate SDC-UDA with multiple publicly available cross-modality medical image segmentation datasets and achieve state-of-the-art segmentation performance, not to mention the superior slice-direction continuity of prediction compared to previous studies.
Evidence-empowered Transfer Learning for Alzheimer's Disease
Mar 03, 2023



Abstract:Transfer learning has been widely utilized to mitigate the data scarcity problem in the field of Alzheimer's disease (AD). Conventional transfer learning relies on re-using models trained on AD-irrelevant tasks such as natural image classification. However, it often leads to negative transfer due to the discrepancy between the non-medical source and target medical domains. To address this, we present evidence-empowered transfer learning for AD diagnosis. Unlike conventional approaches, we leverage an AD-relevant auxiliary task, namely morphological change prediction, without requiring additional MRI data. In this auxiliary task, the diagnosis model learns the evidential and transferable knowledge from morphological features in MRI scans. Experimental results demonstrate that our framework is not only effective in improving detection performance regardless of model capacity, but also more data-efficient and faithful.
COSMOS: Cross-Modality Unsupervised Domain Adaptation for 3D Medical Image Segmentation based on Target-aware Domain Translation and Iterative Self-Training
Mar 30, 2022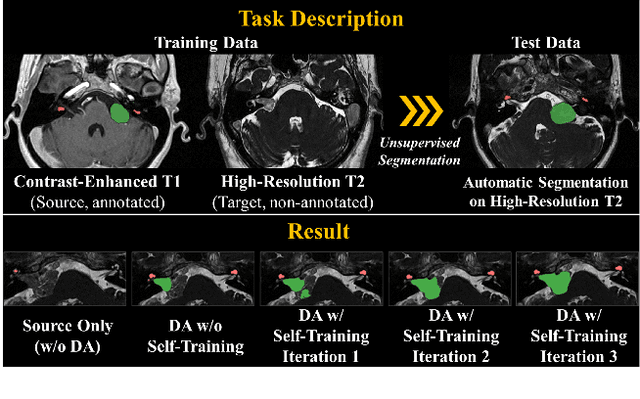
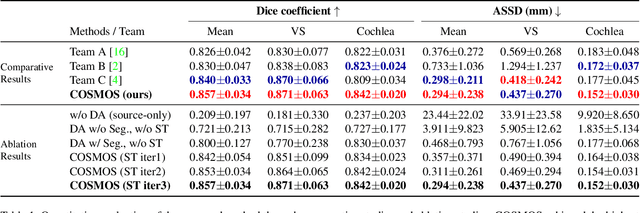
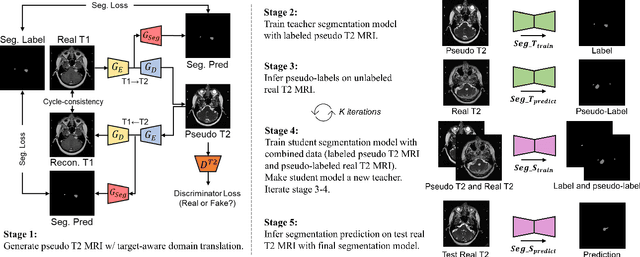
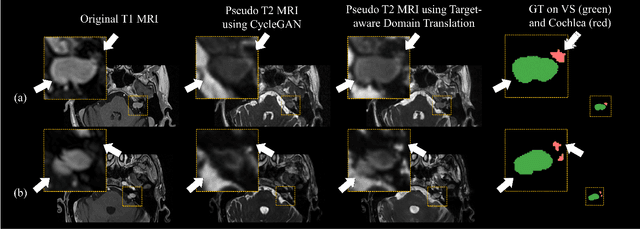
Abstract:Recent advances in deep learning-based medical image segmentation studies achieve nearly human-level performance when in fully supervised condition. However, acquiring pixel-level expert annotations is extremely expensive and laborious in medical imaging fields. Unsupervised domain adaptation can alleviate this problem, which makes it possible to use annotated data in one imaging modality to train a network that can successfully perform segmentation on target imaging modality with no labels. In this work, we propose a self-training based unsupervised domain adaptation framework for 3D medical image segmentation named COSMOS and validate it with automatic segmentation of Vestibular Schwannoma (VS) and cochlea on high-resolution T2 Magnetic Resonance Images (MRI). Our target-aware contrast conversion network translates source domain annotated T1 MRI to pseudo T2 MRI to enable segmentation training on target domain, while preserving important anatomical features of interest in the converted images. Iterative self-training is followed to incorporate unlabeled data to training and incrementally improve the quality of pseudo-labels, thereby leading to improved performance of segmentation. COSMOS won the 1\textsuperscript{st} place in the Cross-Modality Domain Adaptation (crossMoDA) challenge held in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). It achieves mean Dice score and Average Symmetric Surface Distance of 0.871(0.063) and 0.437(0.270) for VS, and 0.842(0.020) and 0.152(0.030) for cochlea.
Self-Training Based Unsupervised Cross-Modality Domain Adaptation for Vestibular Schwannoma and Cochlea Segmentation
Sep 22, 2021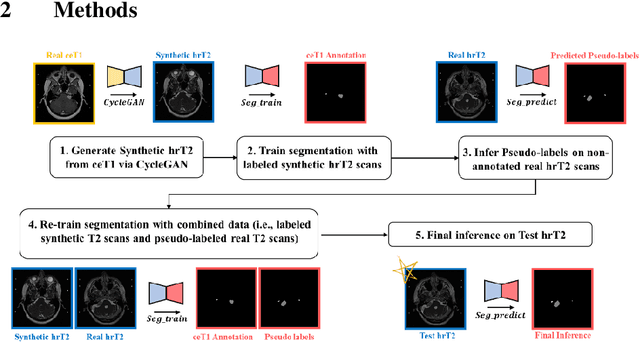
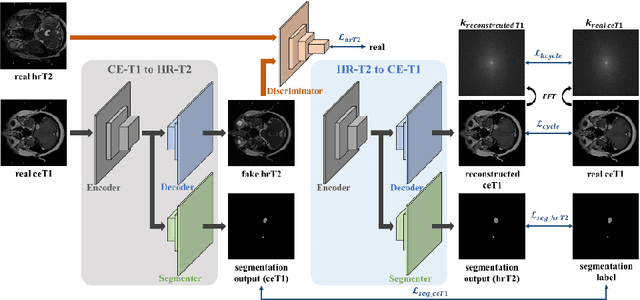
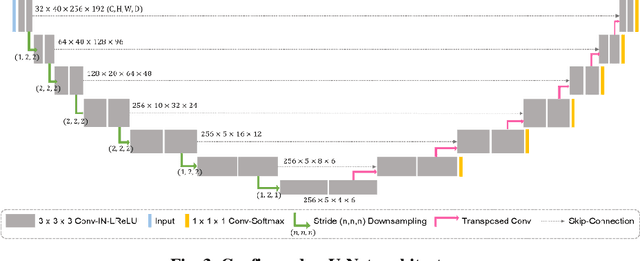
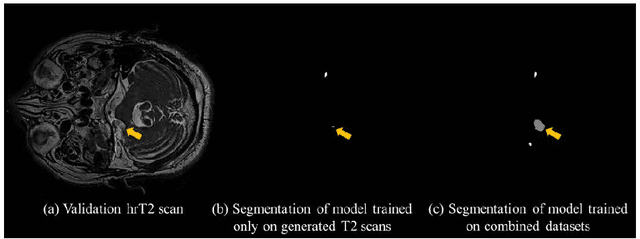
Abstract:With the advances of deep learning, many medical image segmentation studies achieve human-level performance when in fully supervised condition. However, it is extremely expensive to acquire annotation on every data in medical fields, especially on magnetic resonance images (MRI) that comprise many different contrasts. Unsupervised methods can alleviate this problem; however, the performance drop is inevitable compared to fully supervised methods. In this work, we propose a self-training based unsupervised-learning framework that performs automatic segmentation of Vestibular Schwannoma (VS) and cochlea on high-resolution T2 scans. Our method consists of 4 main stages: 1) VS-preserving contrast conversion from contrast-enhanced T1 scan to high-resolution T2 scan, 2) training segmentation on generated T2 scans with annotations on T1 scans, and 3) Inferring pseudo-labels on non-annotated real T2 scans, and 4) boosting the generalizability of VS and cochlea segmentation by training with combined data (i.e., real T2 scans with pseudo-labels and generated T2 scans with true annotations). Our method showed mean Dice score and Average Symmetric Surface Distance (ASSD) of 0.8570 (0.0705) and 0.4970 (0.3391) for VS, 0.8446 (0.0211) and 0.1513 (0.0314) for Cochlea on CrossMoDA2021 challenge validation phase leaderboard, outperforming most other approaches.
State-of-the-Art Machine Learning MRI Reconstruction in 2020: Results of the Second fastMRI Challenge
Dec 28, 2020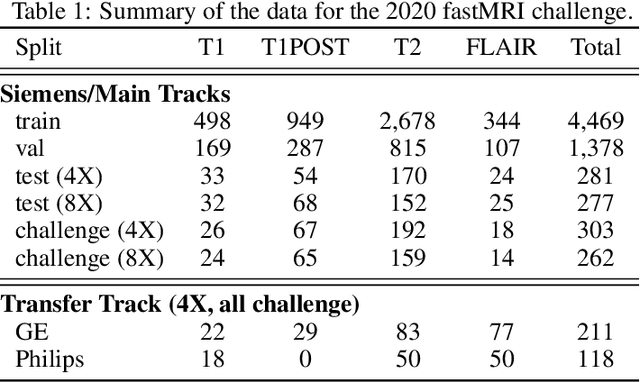
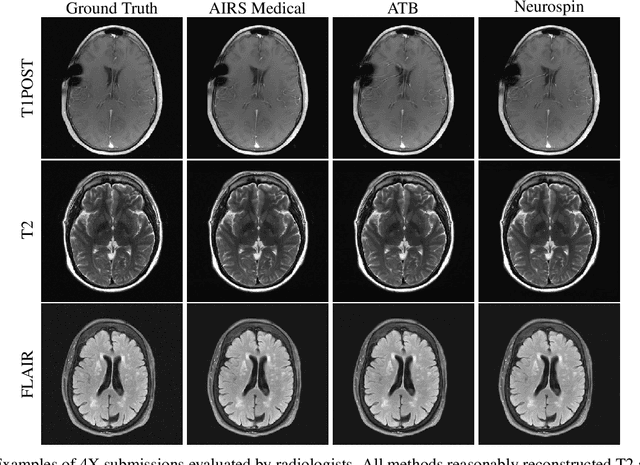
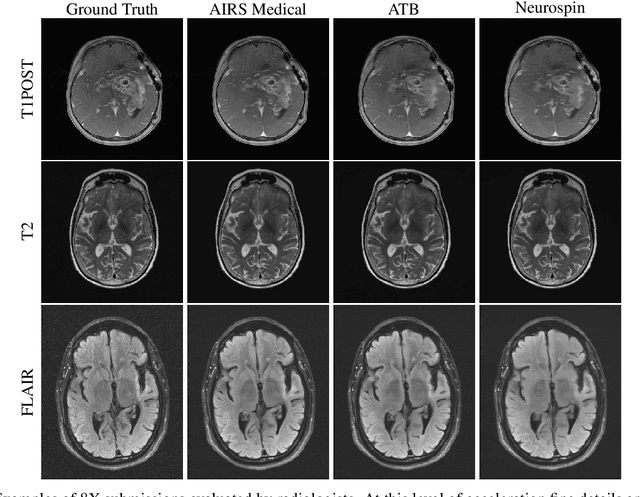
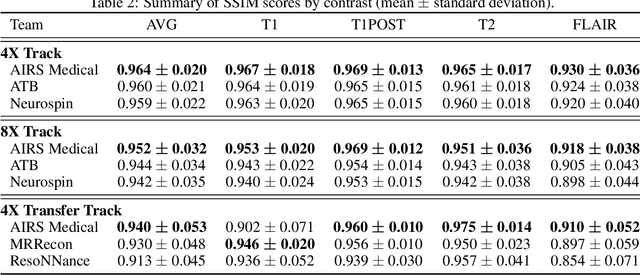
Abstract:Accelerating MRI scans is one of the principal outstanding problems in the MRI research community. Towards this goal, we hosted the second fastMRI competition targeted towards reconstructing MR images with subsampled k-space data. We provided participants with data from 7,299 clinical brain scans (de-identified via a HIPAA-compliant procedure by NYU Langone Health), holding back the fully-sampled data from 894 of these scans for challenge evaluation purposes. In contrast to the 2019 challenge, we focused our radiologist evaluations on pathological assessment in brain images. We also debuted a new Transfer track that required participants to submit models evaluated on MRI scanners from outside the training set. We received 19 submissions from eight different groups. Results showed one team scoring best in both SSIM scores and qualitative radiologist evaluations. We also performed analysis on alternative metrics to mitigate the effects of background noise and collected feedback from the participants to inform future challenges. Lastly, we identify common failure modes across the submissions, highlighting areas of need for future research in the MRI reconstruction community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge