Dingyi Rong
EnerBridge-DPO: Energy-Guided Protein Inverse Folding with Markov Bridges and Direct Preference Optimization
Jun 11, 2025Abstract:Designing protein sequences with optimal energetic stability is a key challenge in protein inverse folding, as current deep learning methods are primarily trained by maximizing sequence recovery rates, often neglecting the energy of the generated sequences. This work aims to overcome this limitation by developing a model that directly generates low-energy, stable protein sequences. We propose EnerBridge-DPO, a novel inverse folding framework focused on generating low-energy, high-stability protein sequences. Our core innovation lies in: First, integrating Markov Bridges with Direct Preference Optimization (DPO), where energy-based preferences are used to fine-tune the Markov Bridge model. The Markov Bridge initiates optimization from an information-rich prior sequence, providing DPO with a pool of structurally plausible sequence candidates. Second, an explicit energy constraint loss is introduced, which enhances the energy-driven nature of DPO based on prior sequences, enabling the model to effectively learn energy representations from a wealth of prior knowledge and directly predict sequence energy values, thereby capturing quantitative features of the energy landscape. Our evaluations demonstrate that EnerBridge-DPO can design protein complex sequences with lower energy while maintaining sequence recovery rates comparable to state-of-the-art models, and accurately predicts $\Delta \Delta G$ values between various sequences.
Autoregressive Enzyme Function Prediction with Multi-scale Multi-modality Fusion
Aug 11, 2024Abstract:Accurate prediction of enzyme function is crucial for elucidating biological mechanisms and driving innovation across various sectors. Existing deep learning methods tend to rely solely on either sequence data or structural data and predict the EC number as a whole, neglecting the intrinsic hierarchical structure of EC numbers. To address these limitations, we introduce MAPred, a novel multi-modality and multi-scale model designed to autoregressively predict the EC number of proteins. MAPred integrates both the primary amino acid sequence and the 3D tokens of proteins, employing a dual-pathway approach to capture comprehensive protein characteristics and essential local functional sites. Additionally, MAPred utilizes an autoregressive prediction network to sequentially predict the digits of the EC number, leveraging the hierarchical organization of EC classifications. Evaluations on benchmark datasets, including New-392, Price, and New-815, demonstrate that our method outperforms existing models, marking a significant advance in the reliability and granularity of protein function prediction within bioinformatics.
GARF:Geometry-Aware Generalized Neural Radiance Field
Dec 07, 2022



Abstract:Neural Radiance Field (NeRF) has revolutionized free viewpoint rendering tasks and achieved impressive results. However, the efficiency and accuracy problems hinder its wide applications. To address these issues, we propose Geometry-Aware Generalized Neural Radiance Field (GARF) with a geometry-aware dynamic sampling (GADS) strategy to perform real-time novel view rendering and unsupervised depth estimation on unseen scenes without per-scene optimization. Distinct from most existing generalized NeRFs, our framework infers the unseen scenes on both pixel-scale and geometry-scale with only a few input images. More specifically, our method learns common attributes of novel-view synthesis by an encoder-decoder structure and a point-level learnable multi-view feature fusion module which helps avoid occlusion. To preserve scene characteristics in the generalized model, we introduce an unsupervised depth estimation module to derive the coarse geometry, narrow down the ray sampling interval to proximity space of the estimated surface and sample in expectation maximum position, constituting Geometry-Aware Dynamic Sampling strategy (GADS). Moreover, we introduce a Multi-level Semantic Consistency loss (MSC) to assist more informative representation learning. Extensive experiments on indoor and outdoor datasets show that comparing with state-of-the-art generalized NeRF methods, GARF reduces samples by more than 25\%, while improving rendering quality and 3D geometry estimation.
Differentiable Projection from Optical Coherence Tomography B-Scan without Retinal Layer Segmentation Supervision
Jun 11, 2022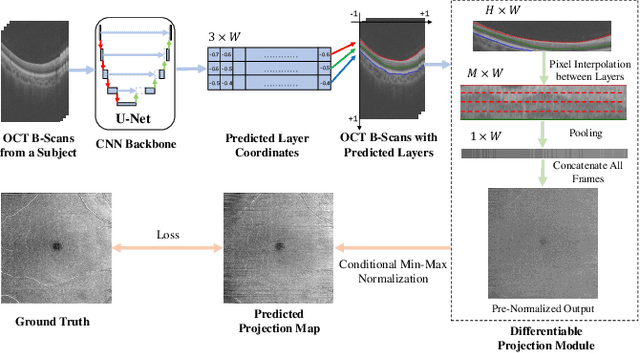
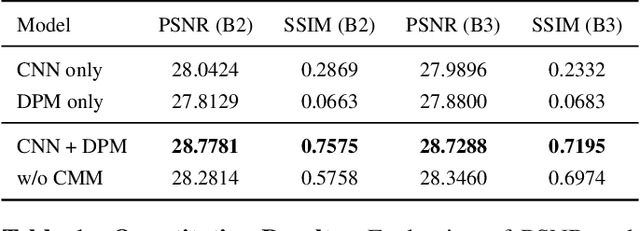
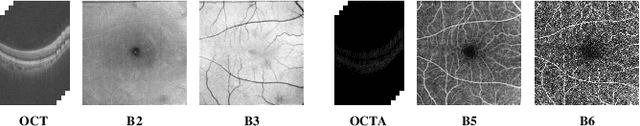
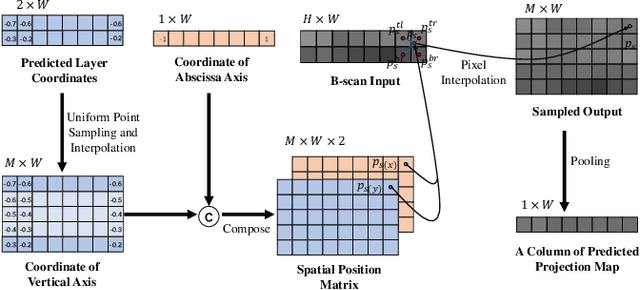
Abstract:Projection map (PM) from optical coherence tomography (OCT) B-scan is an important tool to diagnose retinal diseases, which typically requires retinal layer segmentation. In this study, we present a novel end-to-end framework to predict PMs from B-scans. Instead of segmenting retinal layers explicitly, we represent them implicitly as predicted coordinates. By pixel interpolation on uniformly sampled coordinates between retinal layers, the corresponding PMs could be easily obtained with pooling. Notably, all the operators are differentiable; therefore, this Differentiable Projection Module (DPM) enables end-to-end training with the ground truth of PMs rather than retinal layer segmentation. Our framework produces high-quality PMs, significantly outperforming baselines, including a vanilla CNN without DPM and an optimization-based DPM without a deep prior. Furthermore, the proposed DPM, as a novel neural representation of areas/volumes between curves/surfaces, could be of independent interest for geometric deep learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge