David A. Wolk
for the Alzheimer's Disease Neuroimaging Initiative
Development of an isotropic segmentation model for medial temporal lobe subregions on anisotropic MRI atlas using implicit neural representation
Aug 24, 2025



Abstract:Imaging biomarkers in magnetic resonance imaging (MRI) are important tools for diagnosing and tracking Alzheimer's disease (AD). As medial temporal lobe (MTL) is the earliest region to show AD-related hallmarks, brain atrophy caused by AD can first be observed in the MTL. Accurate segmentation of MTL subregions and extraction of imaging biomarkers from them are important. However, due to imaging limitations, the resolution of T2-weighted (T2w) MRI is anisotropic, which makes it difficult to accurately extract the thickness of cortical subregions in the MTL. In this study, we used an implicit neural representation method to combine the resolution advantages of T1-weighted and T2w MRI to accurately upsample an MTL subregion atlas set from anisotropic space to isotropic space, establishing a multi-modality, high-resolution atlas set. Based on this atlas, we developed an isotropic MTL subregion segmentation model. In an independent test set, the cortical subregion thickness extracted using this isotropic model showed higher significance than an anisotropic method in distinguishing between participants with mild cognitive impairment and cognitively unimpaired (CU) participants. In longitudinal analysis, the biomarkers extracted using isotropic method showed greater stability in CU participants. This study improved the accuracy of AD imaging biomarkers without increasing the amount of atlas annotation work, which may help to more accurately quantify the relationship between AD and brain atrophy and provide more accurate measures for disease tracking.
Nearly isotropic segmentation for medial temporal lobe subregions in multi-modality MRI
Apr 25, 2025Abstract:Morphometry of medial temporal lobe (MTL) subregions in brain MRI is sensitive biomarker to Alzheimers Disease and other related conditions. While T2-weighted (T2w) MRI with high in-plane resolution is widely used to segment hippocampal subfields due to its higher contrast in hippocampus, its lower out-of-plane resolution reduces the accuracy of subregion thickness measurements. To address this issue, we developed a nearly isotropic segmentation pipeline that incorporates image and label upsampling and high-resolution segmentation in T2w MRI. First, a high-resolution atlas was created based on an existing anisotropic atlas derived from 29 individuals. Both T1-weighted and T2w images in the atlas were upsampled from their original resolution to a nearly isotropic resolution 0.4x0.4x0.52mm3 using a non-local means approach. Manual segmentations within the atlas were also upsampled to match this resolution using a UNet-based neural network, which was trained on a cohort consisting of both high-resolution ex vivo and low-resolution anisotropic in vivo MRI with manual segmentations. Second, a multi-modality deep learning-based segmentation model was trained within this nearly isotropic atlas. Finally, experiments showed the nearly isotropic subregion segmentation improved the accuracy of cortical thickness as an imaging biomarker for neurodegeneration in T2w MRI.
Regional Deep Atrophy: a Self-Supervised Learning Method to Automatically Identify Regions Associated With Alzheimer's Disease Progression From Longitudinal MRI
Apr 10, 2023Abstract:Longitudinal assessment of brain atrophy, particularly in the hippocampus, is a well-studied biomarker for neurodegenerative diseases, such as Alzheimer's disease (AD). In clinical trials, estimation of brain progressive rates can be applied to track therapeutic efficacy of disease modifying treatments. However, most state-of-the-art measurements calculate changes directly by segmentation and/or deformable registration of MRI images, and may misreport head motion or MRI artifacts as neurodegeneration, impacting their accuracy. In our previous study, we developed a deep learning method DeepAtrophy that uses a convolutional neural network to quantify differences between longitudinal MRI scan pairs that are associated with time. DeepAtrophy has high accuracy in inferring temporal information from longitudinal MRI scans, such as temporal order or relative inter-scan interval. DeepAtrophy also provides an overall atrophy score that was shown to perform well as a potential biomarker of disease progression and treatment efficacy. However, DeepAtrophy is not interpretable, and it is unclear what changes in the MRI contribute to progression measurements. In this paper, we propose Regional Deep Atrophy (RDA), which combines the temporal inference approach from DeepAtrophy with a deformable registration neural network and attention mechanism that highlights regions in the MRI image where longitudinal changes are contributing to temporal inference. RDA has similar prediction accuracy as DeepAtrophy, but its additional interpretability makes it more acceptable for use in clinical settings, and may lead to more sensitive biomarkers for disease monitoring in clinical trials of early AD.
Automated deep learning segmentation of high-resolution 7 T ex vivo MRI for quantitative analysis of structure-pathology correlations in neurodegenerative diseases
Mar 21, 2023



Abstract:Ex vivo MRI of the brain provides remarkable advantages over in vivo MRI for visualizing and characterizing detailed neuroanatomy, and helps to link microscale histology studies with morphometric measurements. However, automated segmentation methods for brain mapping in ex vivo MRI are not well developed, primarily due to limited availability of labeled datasets, and heterogeneity in scanner hardware and acquisition protocols. In this work, we present a high resolution dataset of 37 ex vivo post-mortem human brain tissue specimens scanned on a 7T whole-body MRI scanner. We developed a deep learning pipeline to segment the cortical mantle by benchmarking the performance of nine deep neural architectures. We then segment the four subcortical structures: caudate, putamen, globus pallidus, and thalamus; white matter hyperintensities, and the normal appearing white matter. We show excellent generalizing capabilities across whole brain hemispheres in different specimens, and also on unseen images acquired at different magnetic field strengths and different imaging sequence. We then compute volumetric and localized cortical thickness measurements across key regions, and link them with semi-quantitative neuropathological ratings. Our code, containerized executables, and the processed datasets are publicly available at: https://github.com/Pulkit-Khandelwal/upenn-picsl-brain-ex-vivo.
Improved Segmentation of Deep Sulci in Cortical Gray Matter Using a Deep Learning Framework Incorporating Laplace's Equation
Mar 03, 2023



Abstract:When developing tools for automated cortical segmentation, the ability to produce topologically correct segmentations is important in order to compute geometrically valid morphometry measures. In practice, accurate cortical segmentation is challenged by image artifacts and the highly convoluted anatomy of the cortex itself. To address this, we propose a novel deep learning-based cortical segmentation method in which prior knowledge about the geometry of the cortex is incorporated into the network during the training process. We design a loss function which uses the theory of Laplace's equation applied to the cortex to locally penalize unresolved boundaries between tightly folded sulci. Using an ex vivo MRI dataset of human medial temporal lobe specimens, we demonstrate that our approach outperforms baseline segmentation networks, both quantitatively and qualitatively.
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023


Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021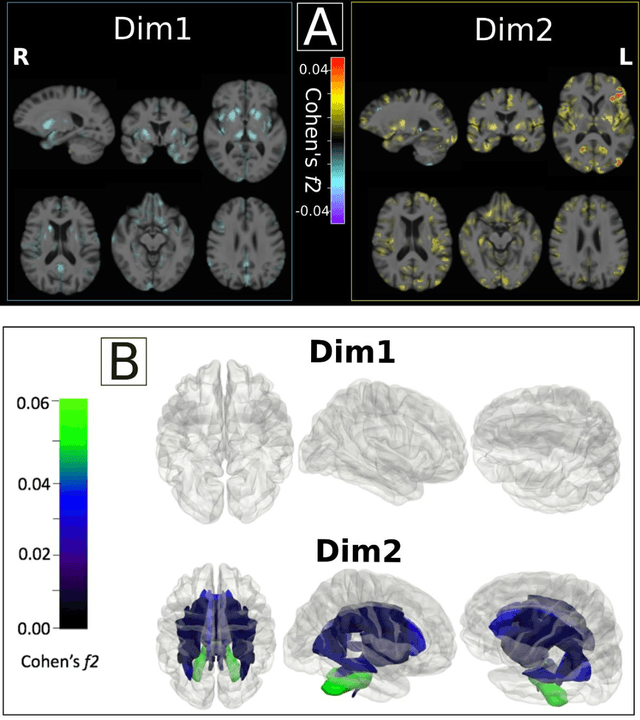
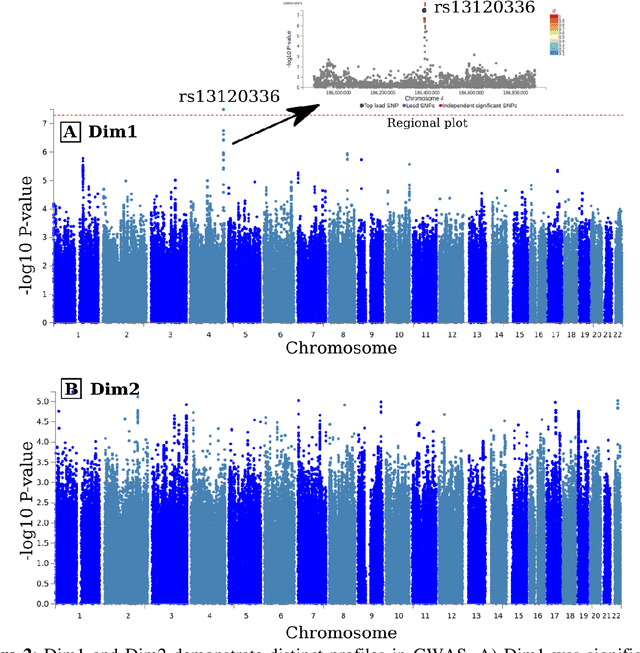
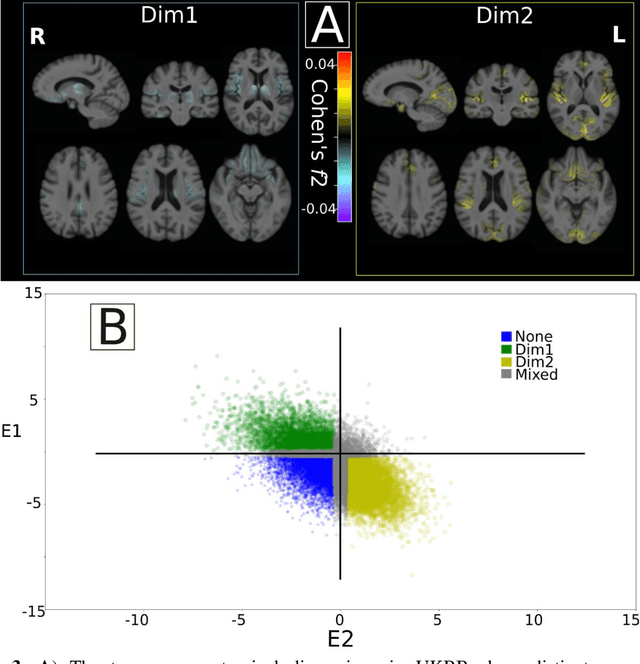
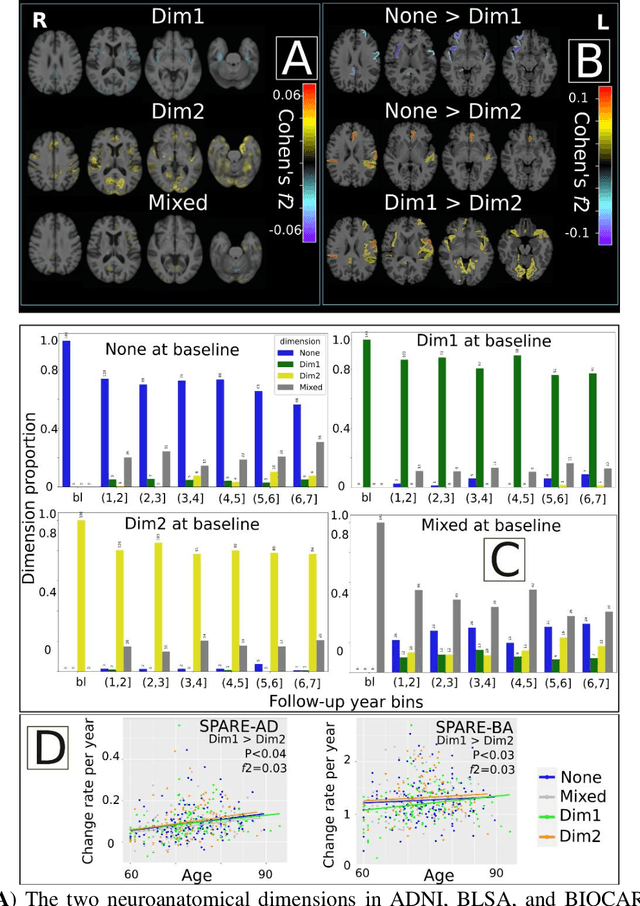
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
Gray Matter Segmentation in Ultra High Resolution 7 Tesla ex vivo T2w MRI of Human Brain Hemispheres
Oct 14, 2021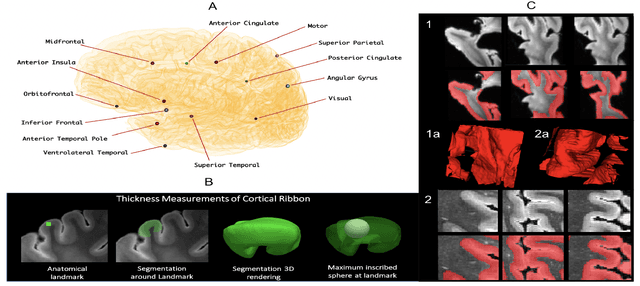
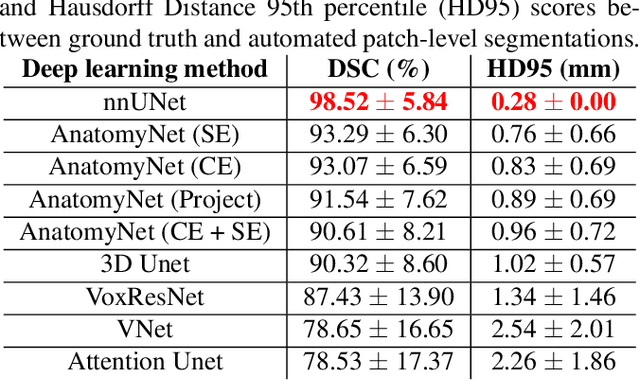
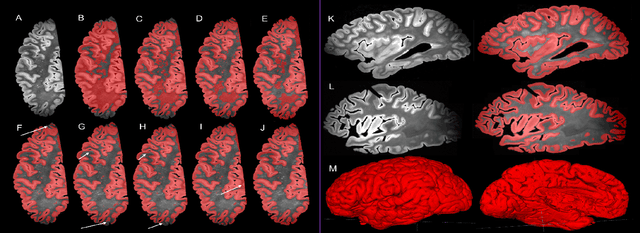
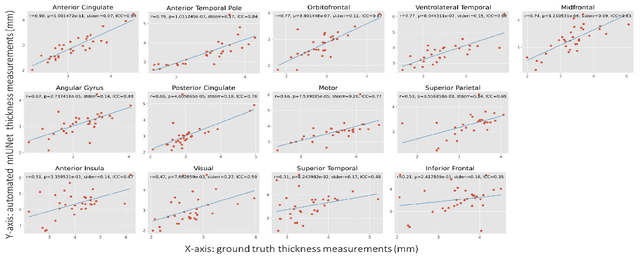
Abstract:Ex vivo MRI of the brain provides remarkable advantages over in vivo MRI for visualizing and characterizing detailed neuroanatomy. However, automated cortical segmentation methods in ex vivo MRI are not well developed, primarily due to limited availability of labeled datasets, and heterogeneity in scanner hardware and acquisition protocols. In this work, we present a high resolution 7 Tesla dataset of 32 ex vivo human brain specimens. We benchmark the cortical mantle segmentation performance of nine neural network architectures, trained and evaluated using manually-segmented 3D patches sampled from specific cortical regions, and show excellent generalizing capabilities across whole brain hemispheres in different specimens, and also on unseen images acquired at different magnetic field strength and imaging sequences. Finally, we provide cortical thickness measurements across key regions in 3D ex vivo human brain images. Our code and processed datasets are publicly available at https://github.com/Pulkit-Khandelwal/picsl-ex-vivo-segmentation.
Disentangling Alzheimer's disease neurodegeneration from typical brain aging using machine learning
Sep 08, 2021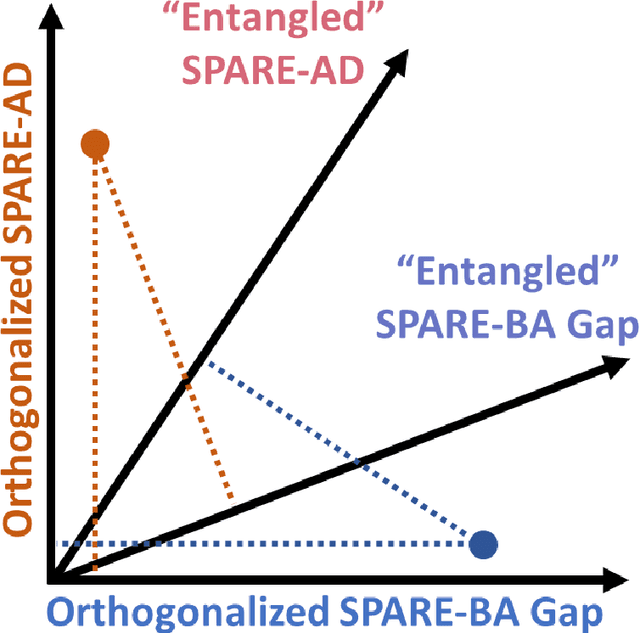
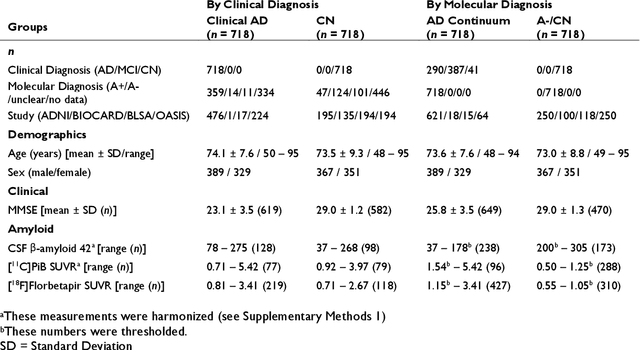
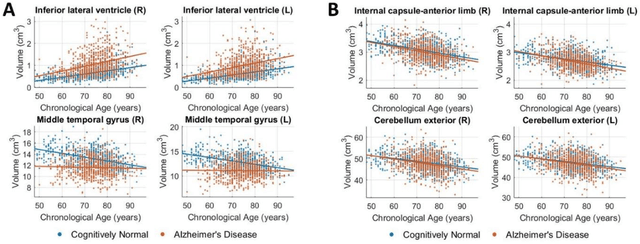
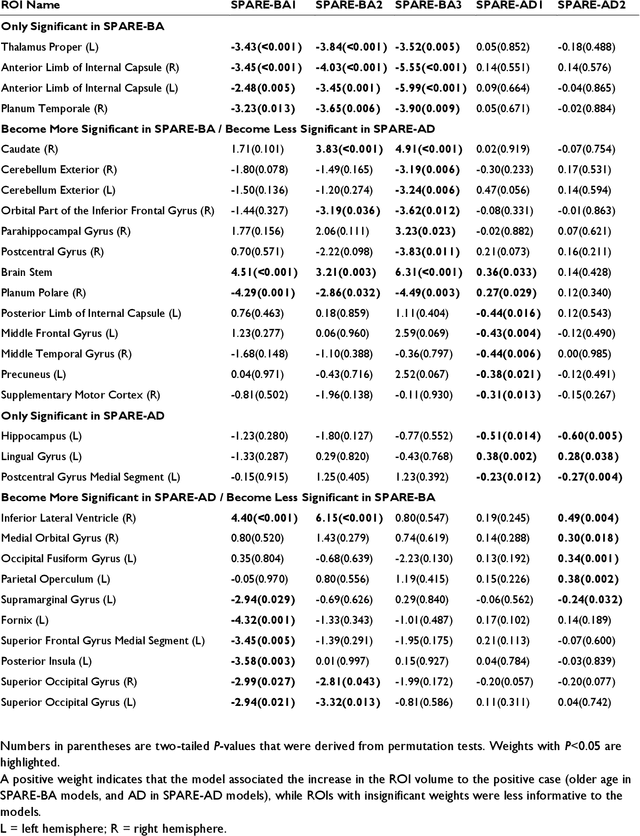
Abstract:Neuroimaging biomarkers that distinguish between typical brain aging and Alzheimer's disease (AD) are valuable for determining how much each contributes to cognitive decline. Machine learning models can derive multi-variate brain change patterns related to the two processes, including the SPARE-AD (Spatial Patterns of Atrophy for Recognition of Alzheimer's Disease) and SPARE-BA (of Brain Aging) investigated herein. However, substantial overlap between brain regions affected in the two processes confounds measuring them independently. We present a methodology toward disentangling the two. T1-weighted MRI images of 4,054 participants (48-95 years) with AD, mild cognitive impairment (MCI), or cognitively normal (CN) diagnoses from the iSTAGING (Imaging-based coordinate SysTem for AGIng and NeurodeGenerative diseases) consortium were analyzed. First, a subset of AD patients and CN adults were selected based purely on clinical diagnoses to train SPARE-BA1 (regression of age using CN individuals) and SPARE-AD1 (classification of CN versus AD). Second, analogous groups were selected based on clinical and molecular markers to train SPARE-BA2 and SPARE-AD2: amyloid-positive (A+) AD continuum group (consisting of A+AD, A+MCI, and A+ and tau-positive CN individuals) and amyloid-negative (A-) CN group. Finally, the combined group of the AD continuum and A-/CN individuals was used to train SPARE-BA3, with the intention to estimate brain age regardless of AD-related brain changes. Disentangled SPARE models derived brain patterns that were more specific to the two types of the brain changes. Correlation between the SPARE-BA and SPARE-AD was significantly reduced. Correlation of disentangled SPARE-AD was non-inferior to the molecular measurements and to the number of APOE4 alleles, but was less to AD-related psychometric test scores, suggesting contribution of advanced brain aging to these scores.
Deep Label Fusion: A 3D End-to-End Hybrid Multi-Atlas Segmentation and Deep Learning Pipeline
Mar 19, 2021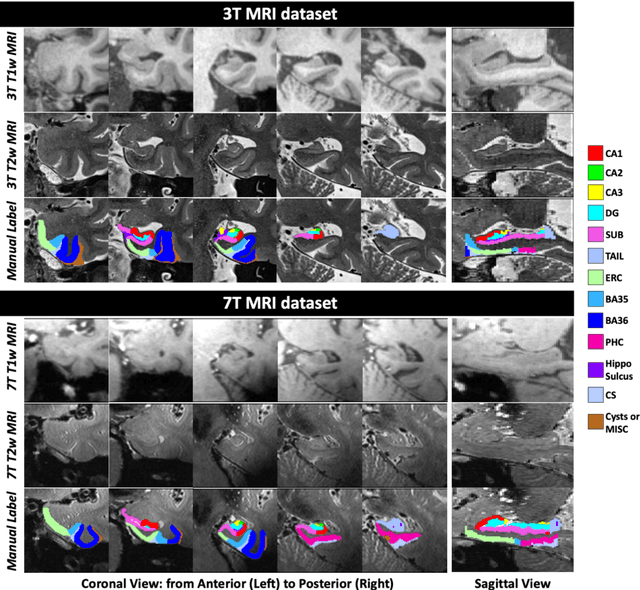
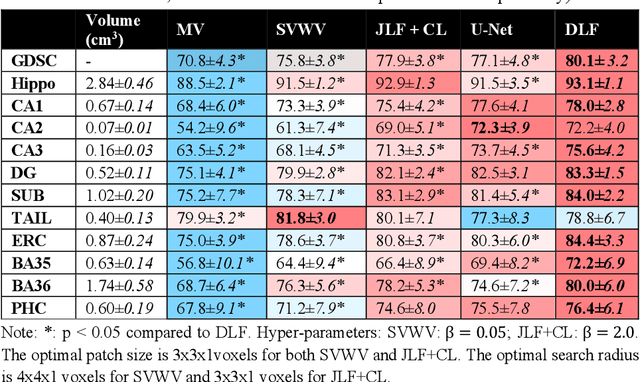
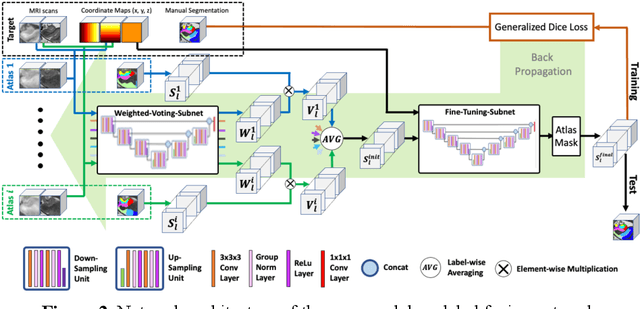
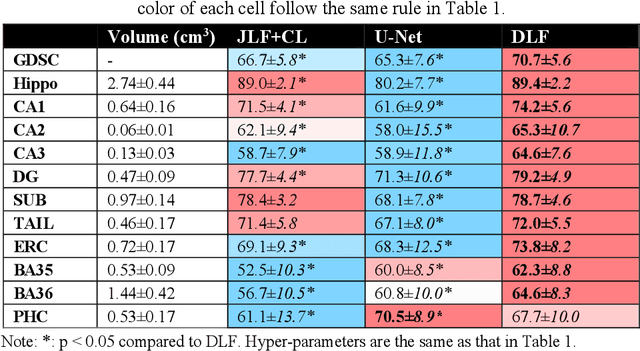
Abstract:Deep learning (DL) is the state-of-the-art methodology in various medical image segmentation tasks. However, it requires relatively large amounts of manually labeled training data, which may be infeasible to generate in some applications. In addition, DL methods have relatively poor generalizability to out-of-sample data. Multi-atlas segmentation (MAS), on the other hand, has promising performance using limited amounts of training data and good generalizability. A hybrid method that integrates the high accuracy of DL and good generalizability of MAS is highly desired and could play an important role in segmentation problems where manually labeled data is hard to generate. Most of the prior work focuses on improving single components of MAS using DL rather than directly optimizing the final segmentation accuracy via an end-to-end pipeline. Only one study explored this idea in binary segmentation of 2D images, but it remains unknown whether it generalizes well to multi-class 3D segmentation problems. In this study, we propose a 3D end-to-end hybrid pipeline, named deep label fusion (DLF), that takes advantage of the strengths of MAS and DL. Experimental results demonstrate that DLF yields significant improvements over conventional label fusion methods and U-Net, a direct DL approach, in the context of segmenting medial temporal lobe subregions using 3T T1-weighted and T2-weighted MRI. Further, when applied to an unseen similar dataset acquired in 7T, DLF maintains its superior performance, which demonstrates its good generalizability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge