Daniel Smilkov
PaLM 2 Technical Report
May 17, 2023



Abstract:We introduce PaLM 2, a new state-of-the-art language model that has better multilingual and reasoning capabilities and is more compute-efficient than its predecessor PaLM. PaLM 2 is a Transformer-based model trained using a mixture of objectives. Through extensive evaluations on English and multilingual language, and reasoning tasks, we demonstrate that PaLM 2 has significantly improved quality on downstream tasks across different model sizes, while simultaneously exhibiting faster and more efficient inference compared to PaLM. This improved efficiency enables broader deployment while also allowing the model to respond faster, for a more natural pace of interaction. PaLM 2 demonstrates robust reasoning capabilities exemplified by large improvements over PaLM on BIG-Bench and other reasoning tasks. PaLM 2 exhibits stable performance on a suite of responsible AI evaluations, and enables inference-time control over toxicity without additional overhead or impact on other capabilities. Overall, PaLM 2 achieves state-of-the-art performance across a diverse set of tasks and capabilities. When discussing the PaLM 2 family, it is important to distinguish between pre-trained models (of various sizes), fine-tuned variants of these models, and the user-facing products that use these models. In particular, user-facing products typically include additional pre- and post-processing steps. Additionally, the underlying models may evolve over time. Therefore, one should not expect the performance of user-facing products to exactly match the results reported in this report.
Similar Image Search for Histopathology: SMILY
Feb 06, 2019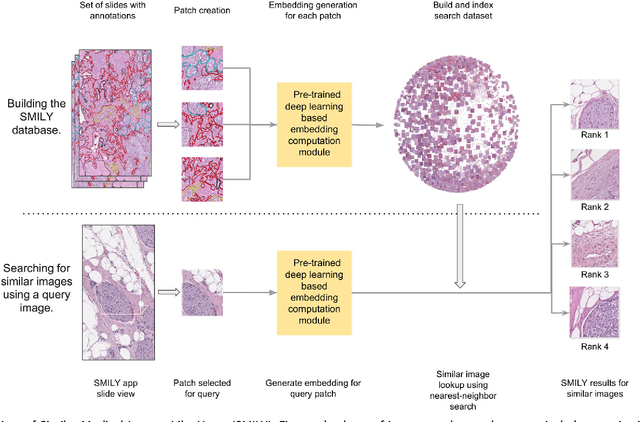
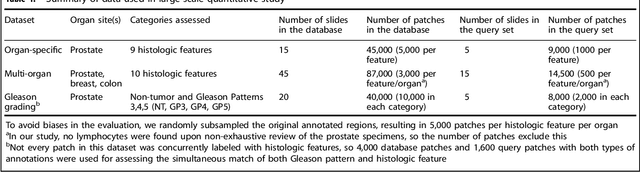
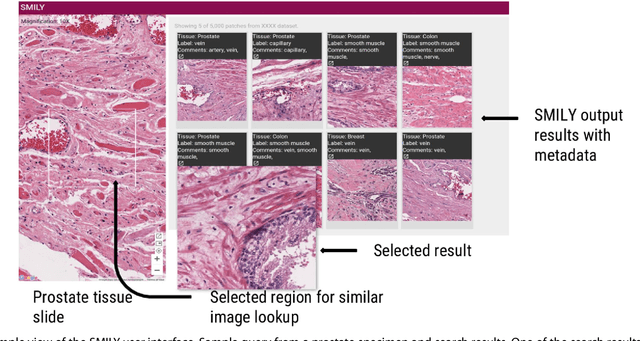
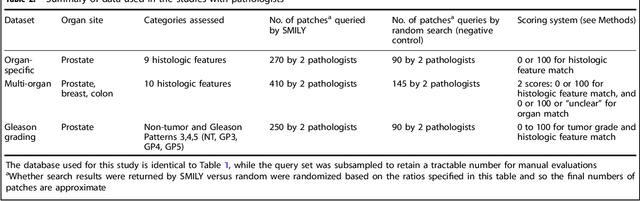
Abstract:The increasing availability of large institutional and public histopathology image datasets is enabling the searching of these datasets for diagnosis, research, and education. Though these datasets typically have associated metadata such as diagnosis or clinical notes, even carefully curated datasets rarely contain annotations of the location of regions of interest on each image. Because pathology images are extremely large (up to 100,000 pixels in each dimension), further laborious visual search of each image may be needed to find the feature of interest. In this paper, we introduce a deep learning based reverse image search tool for histopathology images: Similar Medical Images Like Yours (SMILY). We assessed SMILY's ability to retrieve search results in two ways: using pathologist-provided annotations, and via prospective studies where pathologists evaluated the quality of SMILY search results. As a negative control in the second evaluation, pathologists were blinded to whether search results were retrieved by SMILY or randomly. In both types of assessments, SMILY was able to retrieve search results with similar histologic features, organ site, and prostate cancer Gleason grade compared with the original query. SMILY may be a useful general-purpose tool in the pathologist's arsenal, to improve the efficiency of searching large archives of histopathology images, without the need to develop and implement specific tools for each application.
TensorFlow.js: Machine Learning for the Web and Beyond
Jan 16, 2019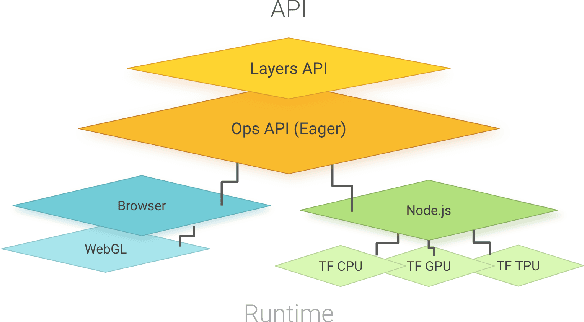
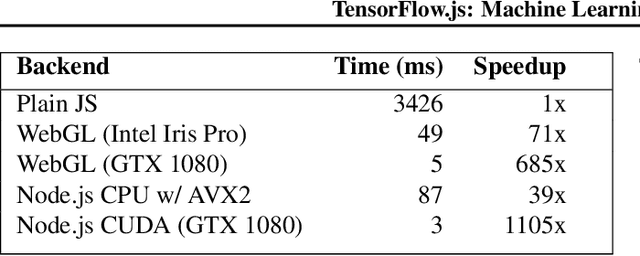
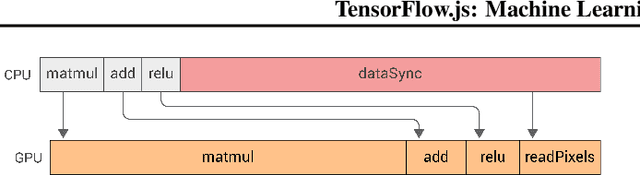

Abstract:TensorFlow.js is a library for building and executing machine learning algorithms in JavaScript. TensorFlow.js models run in a web browser and in the Node.js environment. The library is part of the TensorFlow ecosystem, providing a set of APIs that are compatible with those in Python, allowing models to be ported between the Python and JavaScript ecosystems. TensorFlow.js has empowered a new set of developers from the extensive JavaScript community to build and deploy machine learning models and enabled new classes of on-device computation. This paper describes the design, API, and implementation of TensorFlow.js, and highlights some of the impactful use cases.
Direct-Manipulation Visualization of Deep Networks
Aug 12, 2017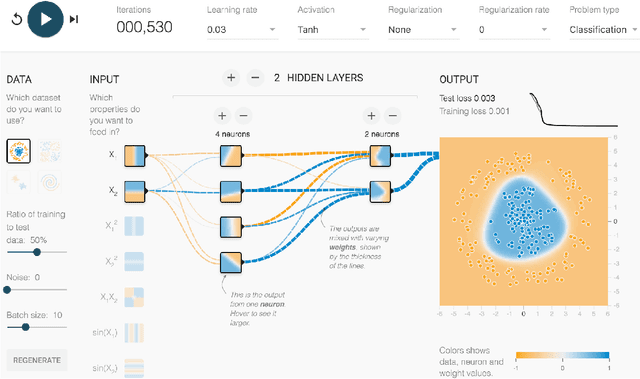
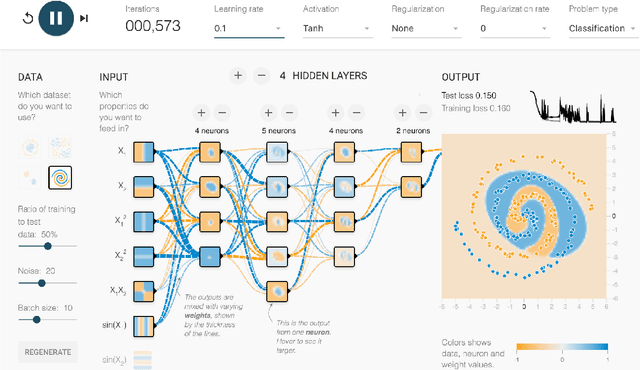
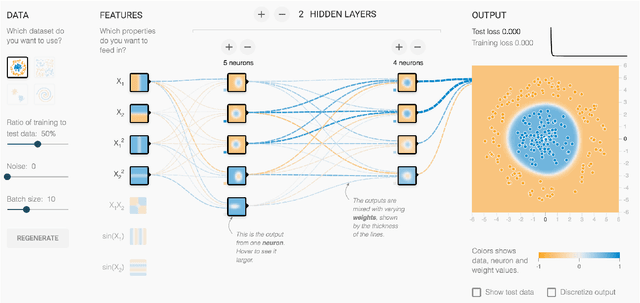
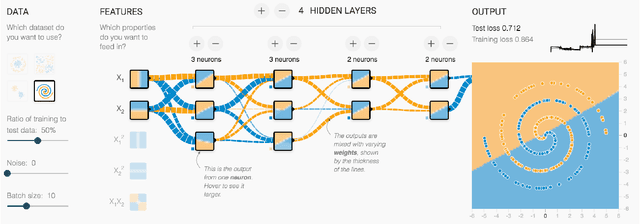
Abstract:The recent successes of deep learning have led to a wave of interest from non-experts. Gaining an understanding of this technology, however, is difficult. While the theory is important, it is also helpful for novices to develop an intuitive feel for the effect of different hyperparameters and structural variations. We describe TensorFlow Playground, an interactive, open sourced visualization that allows users to experiment via direct manipulation rather than coding, enabling them to quickly build an intuition about neural nets.
SmoothGrad: removing noise by adding noise
Jun 12, 2017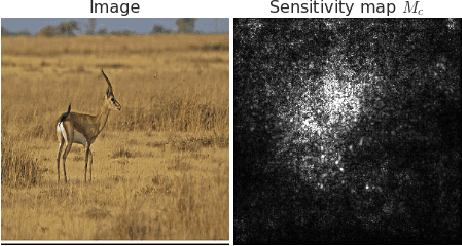
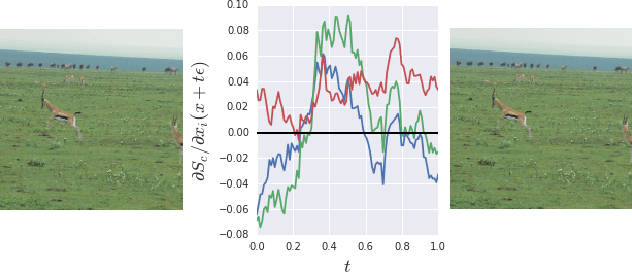
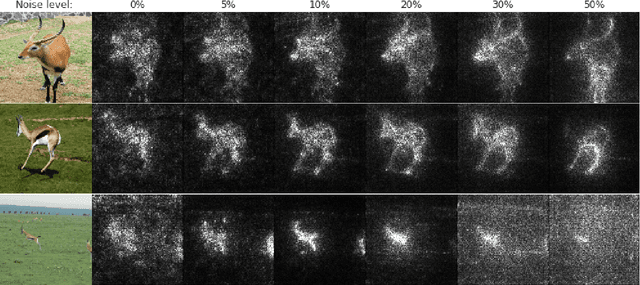

Abstract:Explaining the output of a deep network remains a challenge. In the case of an image classifier, one type of explanation is to identify pixels that strongly influence the final decision. A starting point for this strategy is the gradient of the class score function with respect to the input image. This gradient can be interpreted as a sensitivity map, and there are several techniques that elaborate on this basic idea. This paper makes two contributions: it introduces SmoothGrad, a simple method that can help visually sharpen gradient-based sensitivity maps, and it discusses lessons in the visualization of these maps. We publish the code for our experiments and a website with our results.
Embedding Projector: Interactive Visualization and Interpretation of Embeddings
Nov 16, 2016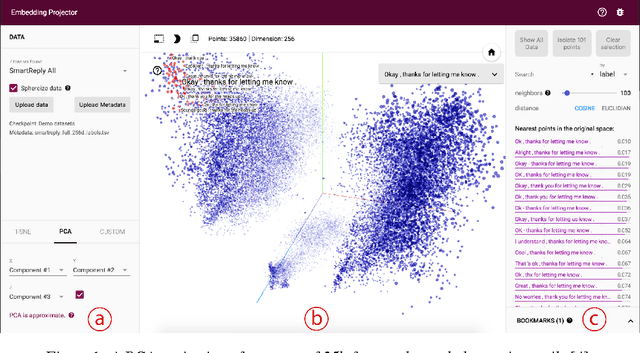
Abstract:Embeddings are ubiquitous in machine learning, appearing in recommender systems, NLP, and many other applications. Researchers and developers often need to explore the properties of a specific embedding, and one way to analyze embeddings is to visualize them. We present the Embedding Projector, a tool for interactive visualization and interpretation of embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge