Bishesh Khanal
Improving Medical Image Classification in Noisy Labels Using Only Self-supervised Pretraining
Aug 08, 2023Abstract:Noisy labels hurt deep learning-based supervised image classification performance as the models may overfit the noise and learn corrupted feature extractors. For natural image classification training with noisy labeled data, model initialization with contrastive self-supervised pretrained weights has shown to reduce feature corruption and improve classification performance. However, no works have explored: i) how other self-supervised approaches, such as pretext task-based pretraining, impact the learning with noisy label, and ii) any self-supervised pretraining methods alone for medical images in noisy label settings. Medical images often feature smaller datasets and subtle inter class variations, requiring human expertise to ensure correct classification. Thus, it is not clear if the methods improving learning with noisy labels in natural image datasets such as CIFAR would also help with medical images. In this work, we explore contrastive and pretext task-based self-supervised pretraining to initialize the weights of a deep learning classification model for two medical datasets with self-induced noisy labels -- NCT-CRC-HE-100K tissue histological images and COVID-QU-Ex chest X-ray images. Our results show that models initialized with pretrained weights obtained from self-supervised learning can effectively learn better features and improve robustness against noisy labels.
M-VAAL: Multimodal Variational Adversarial Active Learning for Downstream Medical Image Analysis Tasks
Jun 21, 2023Abstract:Acquiring properly annotated data is expensive in the medical field as it requires experts, time-consuming protocols, and rigorous validation. Active learning attempts to minimize the need for large annotated samples by actively sampling the most informative examples for annotation. These examples contribute significantly to improving the performance of supervised machine learning models, and thus, active learning can play an essential role in selecting the most appropriate information in deep learning-based diagnosis, clinical assessments, and treatment planning. Although some existing works have proposed methods for sampling the best examples for annotation in medical image analysis, they are not task-agnostic and do not use multimodal auxiliary information in the sampler, which has the potential to increase robustness. Therefore, in this work, we propose a Multimodal Variational Adversarial Active Learning (M-VAAL) method that uses auxiliary information from additional modalities to enhance the active sampling. We applied our method to two datasets: i) brain tumor segmentation and multi-label classification using the BraTS2018 dataset, and ii) chest X-ray image classification using the COVID-QU-Ex dataset. Our results show a promising direction toward data-efficient learning under limited annotations.
Deep-learning assisted detection and quantification of (oo)cysts of Giardia and Cryptosporidium on smartphone microscopy images
Apr 11, 2023Abstract:The consumption of microbial-contaminated food and water is responsible for the deaths of millions of people annually. Smartphone-based microscopy systems are portable, low-cost, and more accessible alternatives for the detection of Giardia and Cryptosporidium than traditional brightfield microscopes. However, the images from smartphone microscopes are noisier and require manual cyst identification by trained technicians, usually unavailable in resource-limited settings. Automatic detection of (oo)cysts using deep-learning-based object detection could offer a solution for this limitation. We evaluate the performance of three state-of-the-art object detectors to detect (oo)cysts of Giardia and Cryptosporidium on a custom dataset that includes both smartphone and brightfield microscopic images from vegetable samples. Faster RCNN, RetinaNet, and you only look once (YOLOv8s) deep-learning models were employed to explore their efficacy and limitations. Our results show that while the deep-learning models perform better with the brightfield microscopy image dataset than the smartphone microscopy image dataset, the smartphone microscopy predictions are still comparable to the prediction performance of non-experts.
COVID-19-related Nepali Tweets Classification in a Low Resource Setting
Oct 11, 2022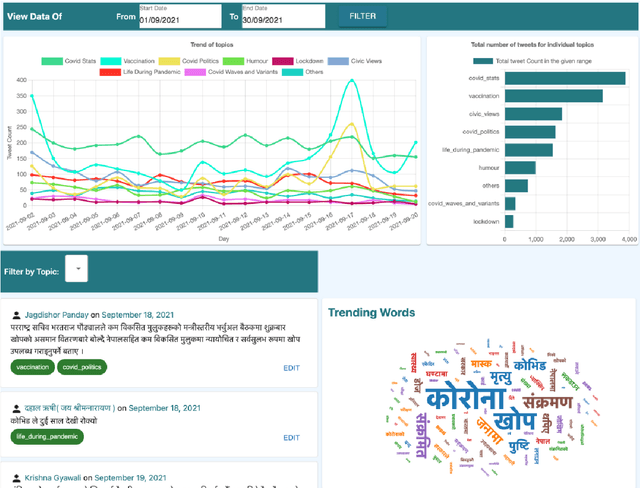
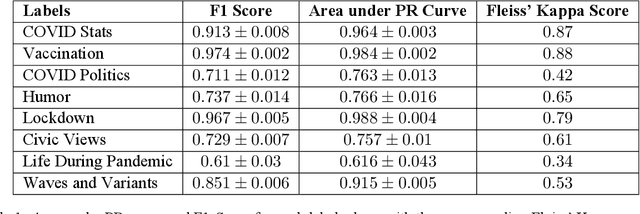
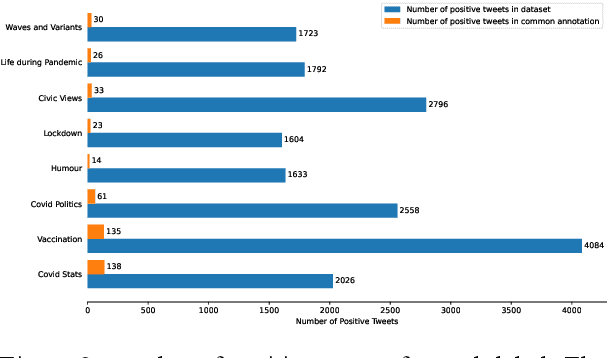
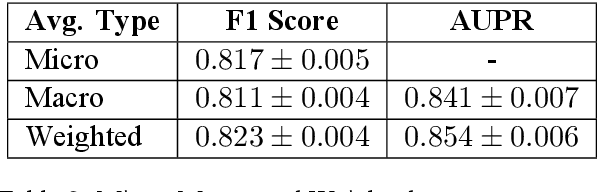
Abstract:Billions of people across the globe have been using social media platforms in their local languages to voice their opinions about the various topics related to the COVID-19 pandemic. Several organizations, including the World Health Organization, have developed automated social media analysis tools that classify COVID-19-related tweets into various topics. However, these tools that help combat the pandemic are limited to very few languages, making several countries unable to take their benefit. While multi-lingual or low-resource language-specific tools are being developed, they still need to expand their coverage, such as for the Nepali language. In this paper, we identify the eight most common COVID-19 discussion topics among the Twitter community using the Nepali language, set up an online platform to automatically gather Nepali tweets containing the COVID-19-related keywords, classify the tweets into the eight topics, and visualize the results across the period in a web-based dashboard. We compare the performance of two state-of-the-art multi-lingual language models for Nepali tweet classification, one generic (mBERT) and the other Nepali language family-specific model (MuRIL). Our results show that the models' relative performance depends on the data size, with MuRIL doing better for a larger dataset. The annotated data, models, and the web-based dashboard are open-sourced at https://github.com/naamiinepal/covid-tweet-classification.
FixMatchSeg: Fixing FixMatch for Semi-Supervised Semantic Segmentation
Aug 02, 2022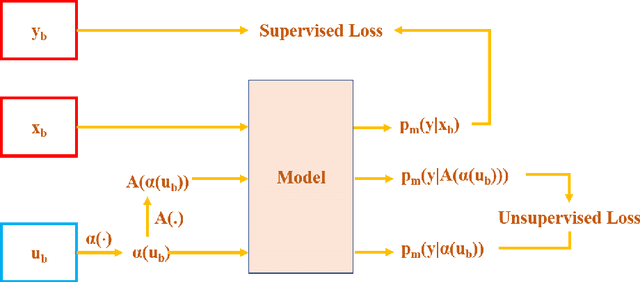
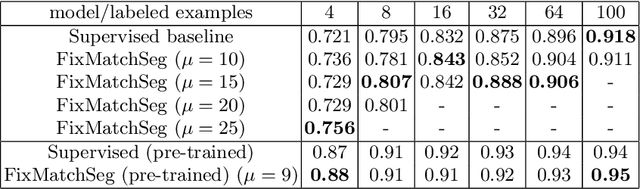
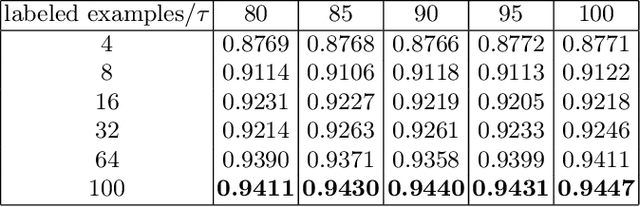
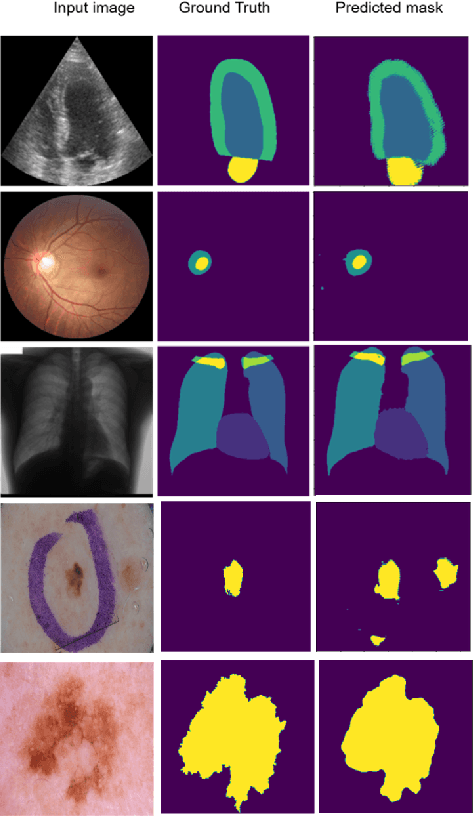
Abstract:Supervised deep learning methods for semantic medical image segmentation are getting increasingly popular in the past few years.However, in resource constrained settings, getting large number of annotated images is very difficult as it mostly requires experts, is expensive and time-consuming.Semi-supervised segmentation can be an attractive solution where a very few labeled images are used along with a large number of unlabeled ones. While the gap between supervised and semi-supervised methods have been dramatically reduced for classification problems in the past couple of years, there still remains a larger gap in segmentation methods. In this work, we adapt a state-of-the-art semi-supervised classification method FixMatch to semantic segmentation task, introducing FixMatchSeg. FixMatchSeg is evaluated in four different publicly available datasets of different anatomy and different modality: cardiac ultrasound, chest X-ray, retinal fundus image, and skin images. When there are few labels, we show that FixMatchSeg performs on par with strong supervised baselines.
Label Geometry Aware Discriminator for Conditional Generative Networks
May 12, 2021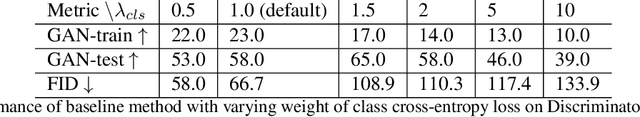
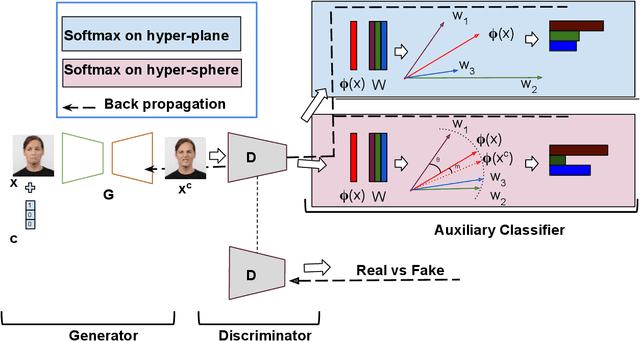
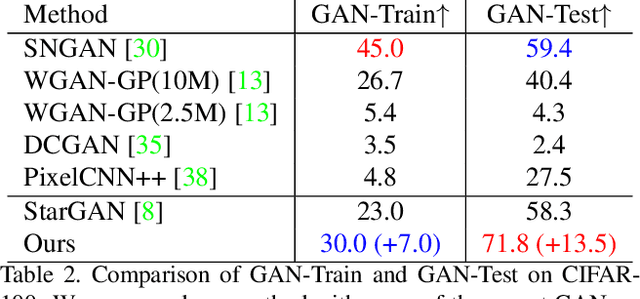
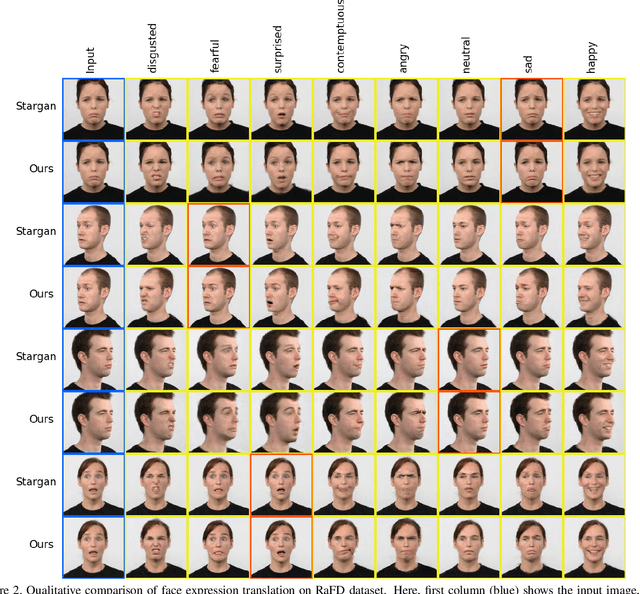
Abstract:Multi-domain image-to-image translation with conditional Generative Adversarial Networks (GANs) can generate highly photo realistic images with desired target classes, yet these synthetic images have not always been helpful to improve downstream supervised tasks such as image classification. Improving downstream tasks with synthetic examples requires generating images with high fidelity to the unknown conditional distribution of the target class, which many labeled conditional GANs attempt to achieve by adding soft-max cross-entropy loss based auxiliary classifier in the discriminator. As recent studies suggest that the soft-max loss in Euclidean space of deep feature does not leverage their intrinsic angular distribution, we propose to replace this loss in auxiliary classifier with an additive angular margin (AAM) loss that takes benefit of the intrinsic angular distribution, and promotes intra-class compactness and inter-class separation to help generator synthesize high fidelity images. We validate our method on RaFD and CIFAR-100, two challenging face expression and natural image classification data set. Our method outperforms state-of-the-art methods in several different evaluation criteria including recently proposed GAN-train and GAN-test metrics designed to assess the impact of synthetic data on downstream classification task, assessing the usefulness in data augmentation for supervised tasks with prediction accuracy score and average confidence score, and the well known FID metric.
Uncertainty Estimation in Deep 2D Echocardiography Segmentation
May 19, 2020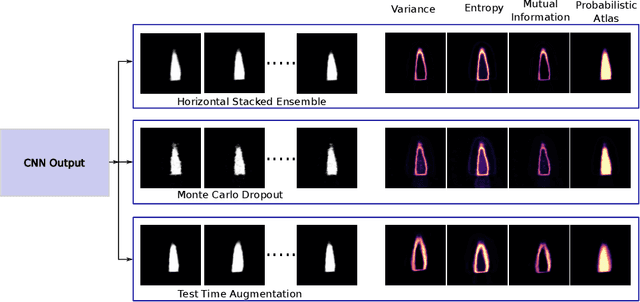
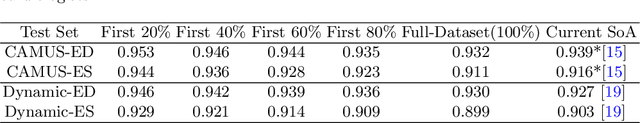
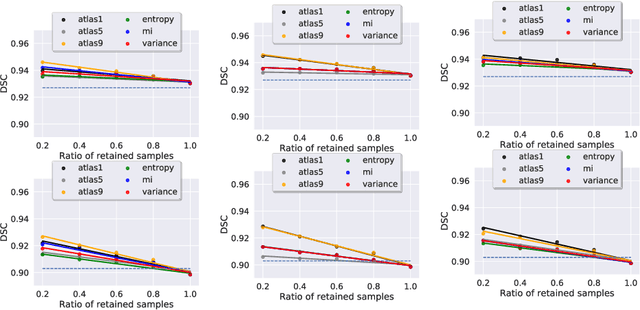
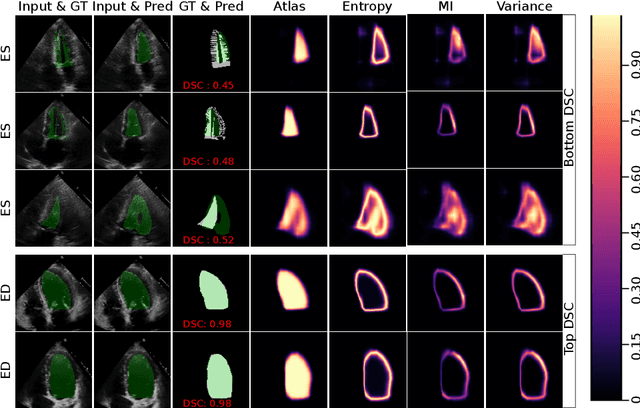
Abstract:2D echocardiography is the most common imaging modality for cardiovascular diseases. The portability and relatively low-cost nature of Ultrasound (US) enable the US devices needed for performing echocardiography to be made widely available. However, acquiring and interpreting cardiac US images is operator dependent, limiting its use to only places where experts are present. Recently, Deep Learning (DL) has been used in 2D echocardiography for automated view classification, and structure and function assessment. Although these recent works show promise in developing computer-guided acquisition and automated interpretation of echocardiograms, most of these methods do not model and estimate uncertainty which can be important when testing on data coming from a distribution further away from that of the training data. Uncertainty estimates can be beneficial both during the image acquisition phase (by providing real-time feedback to the operator on acquired image's quality), and during automated measurement and interpretation. The performance of uncertainty models and quantification metric may depend on the prediction task and the models being compared. Hence, to gain insight of uncertainty modelling for left ventricular segmentation from US images, we compare three ensembling based uncertainty models quantified using four different metrics (one newly proposed) on state-of-the-art baseline networks using two publicly available echocardiogram datasets. We further demonstrate how uncertainty estimation can be used to automatically reject poor quality images and improve state-of-the-art segmentation results.
Automatic Cobb Angle Detection using Vertebra Detector and Vertebra Corners Regression
Oct 31, 2019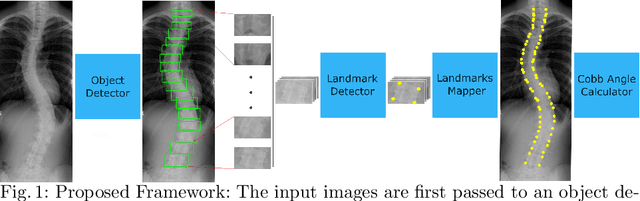

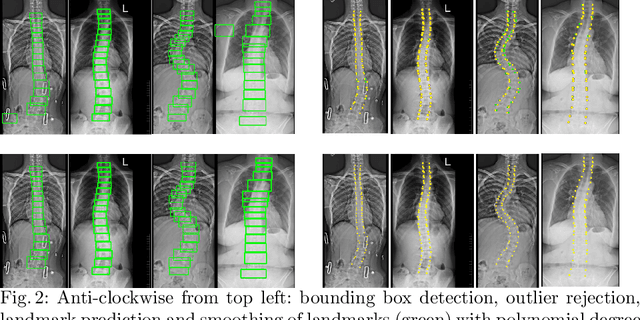
Abstract:Correct evaluation and treatment of Scoliosis require accurate estimation of spinal curvature. Current gold standard is to manually estimate Cobb Angles in spinal X-ray images which is time consuming and has high inter-rater variability. We propose an automatic method with a novel framework that first detects vertebrae as objects followed by a landmark detector that estimates the 4 landmark corners of each vertebra separately. Cobb Angles are calculated using the slope of each vertebra obtained from the predicted landmarks. For inference on test data, we perform pre and post processings that include cropping, outlier rejection and smoothing of the predicted landmarks. The results were assessed in AASCE MICCAI challenge 2019 which showed a promise with a SMAPE score of 25.69 on the challenge test set.
Confident Head Circumference Measurement from Ultrasound with Real-time Feedback for Sonographers
Aug 07, 2019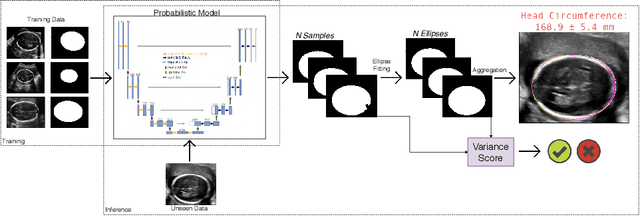
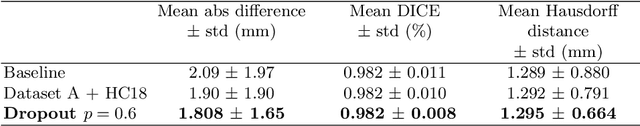
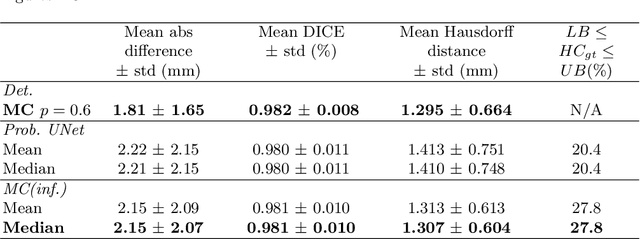
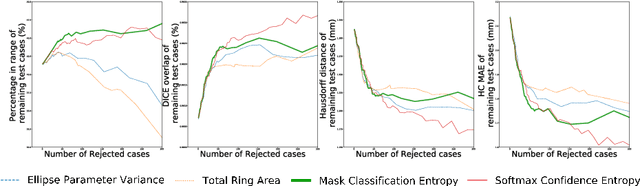
Abstract:Manual estimation of fetal Head Circumference (HC) from Ultrasound (US) is a key biometric for monitoring the healthy development of fetuses. Unfortunately, such measurements are subject to large inter-observer variability, resulting in low early-detection rates of fetal abnormalities. To address this issue, we propose a novel probabilistic Deep Learning approach for real-time automated estimation of fetal HC. This system feeds back statistics on measurement robustness to inform users how confident a deep neural network is in evaluating suitable views acquired during free-hand ultrasound examination. In real-time scenarios, this approach may be exploited to guide operators to scan planes that are as close as possible to the underlying distribution of training images, for the purpose of improving inter-operator consistency. We train on free-hand ultrasound data from over 2000 subjects (2848 training/540 test) and show that our method is able to predict HC measurements within 1.81$\pm$1.65mm deviation from the ground truth, with 50% of the test images fully contained within the predicted confidence margins, and an average of 1.82$\pm$1.78mm deviation from the margin for the remaining cases that are not fully contained.
FastReg: Fast Non-Rigid Registration via Accelerated Optimisation on the Manifold of Diffeomorphisms
Mar 05, 2019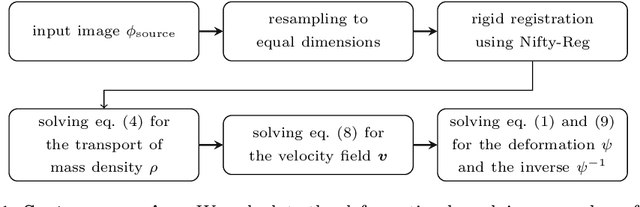
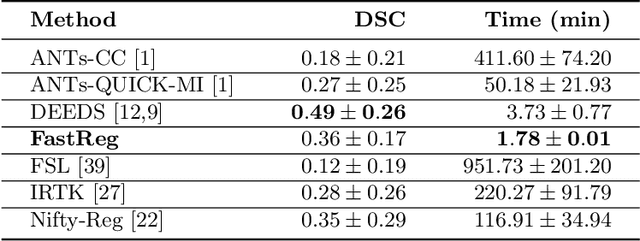
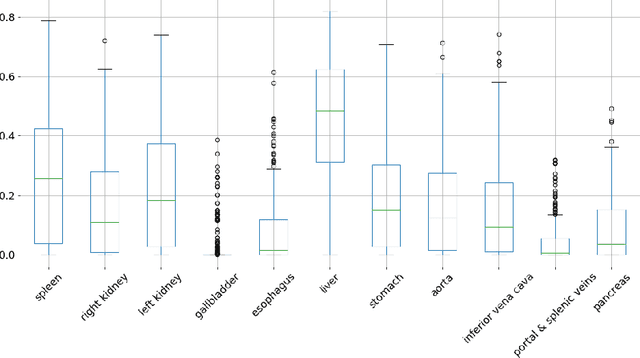
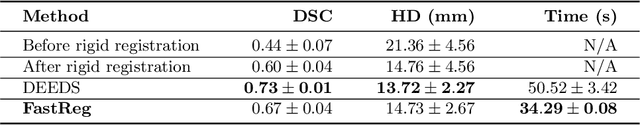
Abstract:We present a new approach to diffeomorphic non-rigid registration of medical images. The method is based on optical flow and warps images via gradient flow with the standard $L^2$ inner product. To compute the transformation, we rely on accelerated optimisation on the manifold of diffeomorphisms. We achieve regularity properties of Sobolev gradient flows, which are expensive to compute, owing to a novel method of averaging the gradients in time rather than space. We successfully register brain MRI and challenging abdominal CT scans at speeds orders of magnitude faster than previous approaches. We make our code available in a public repository: https://github.com/dgrzech/fastreg
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge