Anton Alyakin
Generalist Foundation Models Are Not Clinical Enough for Hospital Operations
Nov 17, 2025Abstract:Hospitals and healthcare systems rely on operational decisions that determine patient flow, cost, and quality of care. Despite strong performance on medical knowledge and conversational benchmarks, foundation models trained on general text may lack the specialized knowledge required for these operational decisions. We introduce Lang1, a family of models (100M-7B parameters) pretrained on a specialized corpus blending 80B clinical tokens from NYU Langone Health's EHRs and 627B tokens from the internet. To rigorously evaluate Lang1 in real-world settings, we developed the REalistic Medical Evaluation (ReMedE), a benchmark derived from 668,331 EHR notes that evaluates five critical tasks: 30-day readmission prediction, 30-day mortality prediction, length of stay, comorbidity coding, and predicting insurance claims denial. In zero-shot settings, both general-purpose and specialized models underperform on four of five tasks (36.6%-71.7% AUROC), with mortality prediction being an exception. After finetuning, Lang1-1B outperforms finetuned generalist models up to 70x larger and zero-shot models up to 671x larger, improving AUROC by 3.64%-6.75% and 1.66%-23.66% respectively. We also observed cross-task scaling with joint finetuning on multiple tasks leading to improvement on other tasks. Lang1-1B effectively transfers to out-of-distribution settings, including other clinical tasks and an external health system. Our findings suggest that predictive capabilities for hospital operations require explicit supervised finetuning, and that this finetuning process is made more efficient by in-domain pretraining on EHR. Our findings support the emerging view that specialized LLMs can compete with generalist models in specialized tasks, and show that effective healthcare systems AI requires the combination of in-domain pretraining, supervised finetuning, and real-world evaluation beyond proxy benchmarks.
Evaluating the performance and fragility of large language models on the self-assessment for neurological surgeons
May 29, 2025


Abstract:The Congress of Neurological Surgeons Self-Assessment for Neurological Surgeons (CNS-SANS) questions are widely used by neurosurgical residents to prepare for written board examinations. Recently, these questions have also served as benchmarks for evaluating large language models' (LLMs) neurosurgical knowledge. This study aims to assess the performance of state-of-the-art LLMs on neurosurgery board-like questions and to evaluate their robustness to the inclusion of distractor statements. A comprehensive evaluation was conducted using 28 large language models. These models were tested on 2,904 neurosurgery board examination questions derived from the CNS-SANS. Additionally, the study introduced a distraction framework to assess the fragility of these models. The framework incorporated simple, irrelevant distractor statements containing polysemous words with clinical meanings used in non-clinical contexts to determine the extent to which such distractions degrade model performance on standard medical benchmarks. 6 of the 28 tested LLMs achieved board-passing outcomes, with the top-performing models scoring over 15.7% above the passing threshold. When exposed to distractions, accuracy across various model architectures was significantly reduced-by as much as 20.4%-with one model failing that had previously passed. Both general-purpose and medical open-source models experienced greater performance declines compared to proprietary variants when subjected to the added distractors. While current LLMs demonstrate an impressive ability to answer neurosurgery board-like exam questions, their performance is markedly vulnerable to extraneous, distracting information. These findings underscore the critical need for developing novel mitigation strategies aimed at bolstering LLM resilience against in-text distractions, particularly for safe and effective clinical deployment.
Gateformer: Advancing Multivariate Time Series Forecasting through Temporal and Variate-Wise Attention with Gated Representations
May 01, 2025Abstract:There has been a recent surge of interest in time series modeling using the Transformer architecture. However, forecasting multivariate time series with Transformer presents a unique challenge as it requires modeling both temporal (cross-time) and variate (cross-variate) dependencies. While Transformer-based models have gained popularity for their flexibility in capturing both sequential and cross-variate relationships, it is unclear how to best integrate these two sources of information in the context of the Transformer architecture while optimizing for both performance and efficiency. We re-purpose the Transformer architecture to effectively model both cross-time and cross-variate dependencies. Our approach begins by embedding each variate independently into a variate-wise representation that captures its cross-time dynamics, and then models cross-variate dependencies through attention mechanisms on these learned embeddings. Gating operations in both cross-time and cross-variate modeling phases regulate information flow, allowing the model to focus on the most relevant features for accurate predictions. Our method achieves state-of-the-art performance across 13 real-world datasets and can be seamlessly integrated into other Transformer-based and LLM-based forecasters, delivering performance improvements up to 20.7\% over original models. Code is available at this repository: https://github.com/nyuolab/Gateformer.
Medical large language models are easily distracted
Apr 01, 2025Abstract:Large language models (LLMs) have the potential to transform medicine, but real-world clinical scenarios contain extraneous information that can hinder performance. The rise of assistive technologies like ambient dictation, which automatically generates draft notes from live patient encounters, has the potential to introduce additional noise making it crucial to assess the ability of LLM's to filter relevant data. To investigate this, we developed MedDistractQA, a benchmark using USMLE-style questions embedded with simulated real-world distractions. Our findings show that distracting statements (polysemous words with clinical meanings used in a non-clinical context or references to unrelated health conditions) can reduce LLM accuracy by up to 17.9%. Commonly proposed solutions to improve model performance such as retrieval-augmented generation (RAG) and medical fine-tuning did not change this effect and in some cases introduced their own confounders and further degraded performance. Our findings suggest that LLMs natively lack the logical mechanisms necessary to distinguish relevant from irrelevant clinical information, posing challenges for real-world applications. MedDistractQA and our results highlights the need for robust mitigation strategies to enhance LLM resilience to extraneous information.
BPQA Dataset: Evaluating How Well Language Models Leverage Blood Pressures to Answer Biomedical Questions
Mar 06, 2025Abstract:Clinical measurements such as blood pressures and respiration rates are critical in diagnosing and monitoring patient outcomes. It is an important component of biomedical data, which can be used to train transformer-based language models (LMs) for improving healthcare delivery. It is, however, unclear whether LMs can effectively interpret and use clinical measurements. We investigate two questions: First, can LMs effectively leverage clinical measurements to answer related medical questions? Second, how to enhance an LM's performance on medical question-answering (QA) tasks that involve measurements? We performed a case study on blood pressure readings (BPs), a vital sign routinely monitored by medical professionals. We evaluated the performance of four LMs: BERT, BioBERT, MedAlpaca, and GPT-3.5, on our newly developed dataset, BPQA (Blood Pressure Question Answering). BPQA contains $100$ medical QA pairs that were verified by medical students and designed to rely on BPs . We found that GPT-3.5 and MedAlpaca (larger and medium sized LMs) benefit more from the inclusion of BPs than BERT and BioBERT (small sized LMs). Further, augmenting measurements with labels improves the performance of BioBERT and Medalpaca (domain specific LMs), suggesting that retrieval may be useful for improving domain-specific LMs.
Repurposing the scientific literature with vision-language models
Feb 26, 2025Abstract:Research in AI for Science often focuses on using AI technologies to augment components of the scientific process, or in some cases, the entire scientific method; how about AI for scientific publications? Peer-reviewed journals are foundational repositories of specialized knowledge, written in discipline-specific language that differs from general Internet content used to train most large language models (LLMs) and vision-language models (VLMs). We hypothesized that by combining a family of scientific journals with generative AI models, we could invent novel tools for scientific communication, education, and clinical care. We converted 23,000 articles from Neurosurgery Publications into a multimodal database - NeuroPubs - of 134 million words and 78,000 image-caption pairs to develop six datasets for building AI models. We showed that the content of NeuroPubs uniquely represents neurosurgery-specific clinical contexts compared with broader datasets and PubMed. For publishing, we employed generalist VLMs to automatically generate graphical abstracts from articles. Editorial board members rated 70% of these as ready for publication without further edits. For education, we generated 89,587 test questions in the style of the ABNS written board exam, which trainee and faculty neurosurgeons found indistinguishable from genuine examples 54% of the time. We used these questions alongside a curriculum learning process to track knowledge acquisition while training our 34 billion-parameter VLM (CNS-Obsidian). In a blinded, randomized controlled trial, we demonstrated the non-inferiority of CNS-Obsidian to GPT-4o (p = 0.1154) as a diagnostic copilot for a neurosurgical service. Our findings lay a novel foundation for AI with Science and establish a framework to elevate scientific communication using state-of-the-art generative artificial intelligence while maintaining rigorous quality standards.
MedG-KRP: Medical Graph Knowledge Representation Probing
Dec 17, 2024Abstract:Large language models (LLMs) have recently emerged as powerful tools, finding many medical applications. LLMs' ability to coalesce vast amounts of information from many sources to generate a response-a process similar to that of a human expert-has led many to see potential in deploying LLMs for clinical use. However, medicine is a setting where accurate reasoning is paramount. Many researchers are questioning the effectiveness of multiple choice question answering (MCQA) benchmarks, frequently used to test LLMs. Researchers and clinicians alike must have complete confidence in LLMs' abilities for them to be deployed in a medical setting. To address this need for understanding, we introduce a knowledge graph (KG)-based method to evaluate the biomedical reasoning abilities of LLMs. Essentially, we map how LLMs link medical concepts in order to better understand how they reason. We test GPT-4, Llama3-70b, and PalmyraMed-70b, a specialized medical model. We enlist a panel of medical students to review a total of 60 LLM-generated graphs and compare these graphs to BIOS, a large biomedical KG. We observe GPT-4 to perform best in our human review but worst in our ground truth comparison; vice-versa with PalmyraMed, the medical model. Our work provides a means of visualizing the medical reasoning pathways of LLMs so they can be implemented in clinical settings safely and effectively.
An Analysis of Euclidean vs. Graph-Based Framing for Bilingual Lexicon Induction from Word Embedding Spaces
Sep 26, 2021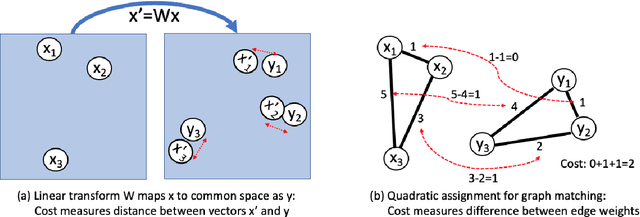

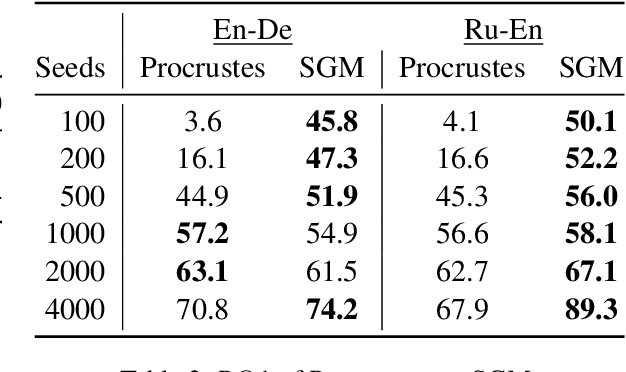
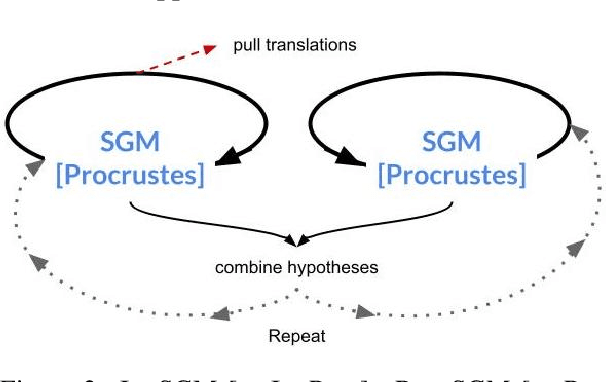
Abstract:Much recent work in bilingual lexicon induction (BLI) views word embeddings as vectors in Euclidean space. As such, BLI is typically solved by finding a linear transformation that maps embeddings to a common space. Alternatively, word embeddings may be understood as nodes in a weighted graph. This framing allows us to examine a node's graph neighborhood without assuming a linear transform, and exploits new techniques from the graph matching optimization literature. These contrasting approaches have not been compared in BLI so far. In this work, we study the behavior of Euclidean versus graph-based approaches to BLI under differing data conditions and show that they complement each other when combined. We release our code at https://github.com/kellymarchisio/euc-v-graph-bli.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge