Mustafa Nasir-Moin
MedG-KRP: Medical Graph Knowledge Representation Probing
Dec 17, 2024Abstract:Large language models (LLMs) have recently emerged as powerful tools, finding many medical applications. LLMs' ability to coalesce vast amounts of information from many sources to generate a response-a process similar to that of a human expert-has led many to see potential in deploying LLMs for clinical use. However, medicine is a setting where accurate reasoning is paramount. Many researchers are questioning the effectiveness of multiple choice question answering (MCQA) benchmarks, frequently used to test LLMs. Researchers and clinicians alike must have complete confidence in LLMs' abilities for them to be deployed in a medical setting. To address this need for understanding, we introduce a knowledge graph (KG)-based method to evaluate the biomedical reasoning abilities of LLMs. Essentially, we map how LLMs link medical concepts in order to better understand how they reason. We test GPT-4, Llama3-70b, and PalmyraMed-70b, a specialized medical model. We enlist a panel of medical students to review a total of 60 LLM-generated graphs and compare these graphs to BIOS, a large biomedical KG. We observe GPT-4 to perform best in our human review but worst in our ground truth comparison; vice-versa with PalmyraMed, the medical model. Our work provides a means of visualizing the medical reasoning pathways of LLMs so they can be implemented in clinical settings safely and effectively.
Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging
Mar 23, 2023Abstract:Molecular classification has transformed the management of brain tumors by enabling more accurate prognostication and personalized treatment. However, timely molecular diagnostic testing for patients with brain tumors is limited, complicating surgical and adjuvant treatment and obstructing clinical trial enrollment. In this study, we developed DeepGlioma, a rapid ($< 90$ seconds), artificial-intelligence-based diagnostic screening system to streamline the molecular diagnosis of diffuse gliomas. DeepGlioma is trained using a multimodal dataset that includes stimulated Raman histology (SRH); a rapid, label-free, non-consumptive, optical imaging method; and large-scale, public genomic data. In a prospective, multicenter, international testing cohort of patients with diffuse glioma ($n=153$) who underwent real-time SRH imaging, we demonstrate that DeepGlioma can predict the molecular alterations used by the World Health Organization to define the adult-type diffuse glioma taxonomy (IDH mutation, 1p19q co-deletion and ATRX mutation), achieving a mean molecular classification accuracy of $93.3\pm 1.6\%$. Our results represent how artificial intelligence and optical histology can be used to provide a rapid and scalable adjunct to wet lab methods for the molecular screening of patients with diffuse glioma.
Identifying and mitigating bias in algorithms used to manage patients in a pandemic
Oct 30, 2021
Abstract:Numerous COVID-19 clinical decision support systems have been developed. However many of these systems do not have the merit for validity due to methodological shortcomings including algorithmic bias. Methods Logistic regression models were created to predict COVID-19 mortality, ventilator status and inpatient status using a real-world dataset consisting of four hospitals in New York City and analyzed for biases against race, gender and age. Simple thresholding adjustments were applied in the training process to establish more equitable models. Results Compared to the naively trained models, the calibrated models showed a 57% decrease in the number of biased trials, while predictive performance, measured by area under the receiver/operating curve (AUC), remained unchanged. After calibration, the average sensitivity of the predictive models increased from 0.527 to 0.955. Conclusion We demonstrate that naively training and deploying machine learning models on real world data for predictive analytics of COVID-19 has a high risk of bias. Simple implemented adjustments or calibrations during model training can lead to substantial and sustained gains in fairness on subsequent deployment.
Patient level simulation and reinforcement learning to discover novel strategies for treating ovarian cancer
Oct 22, 2021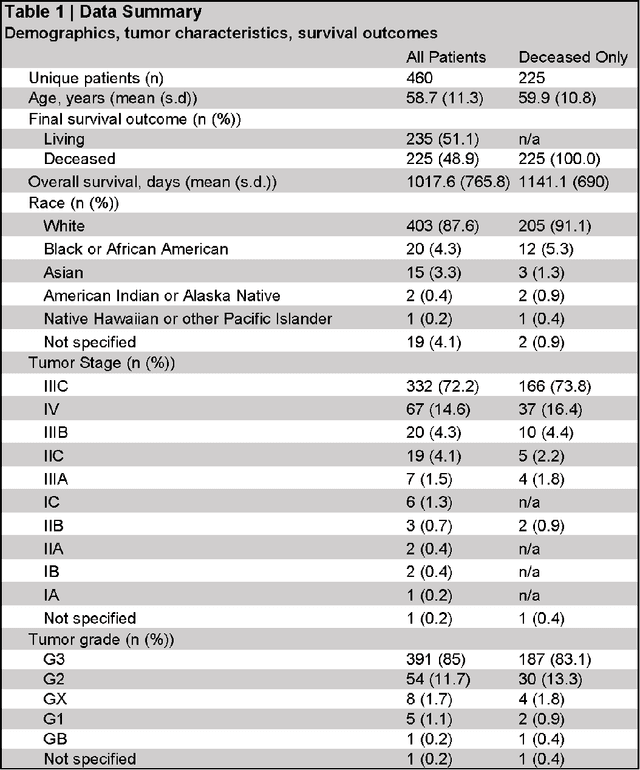
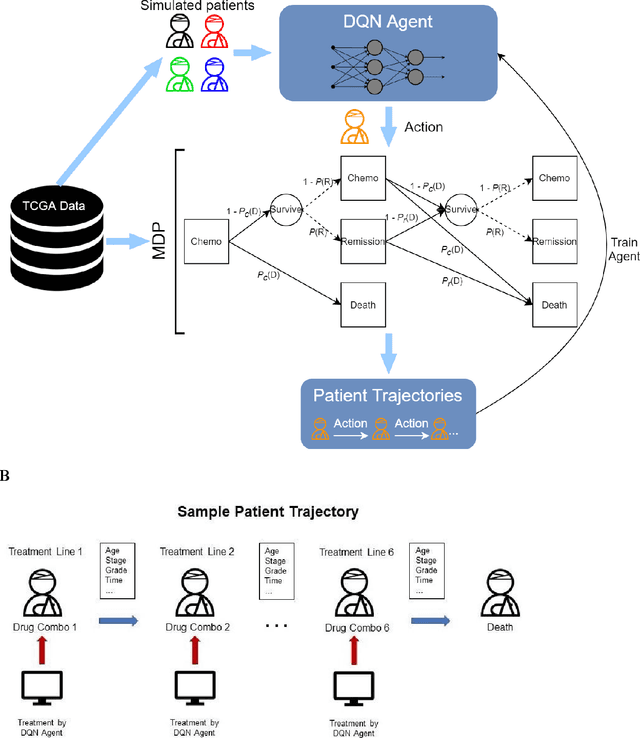
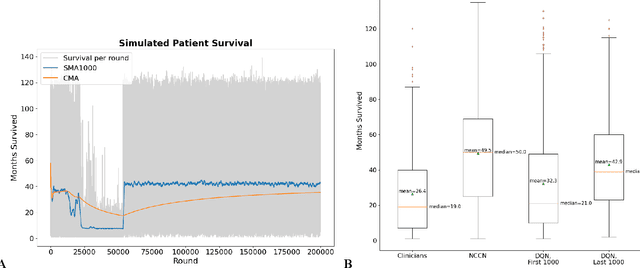
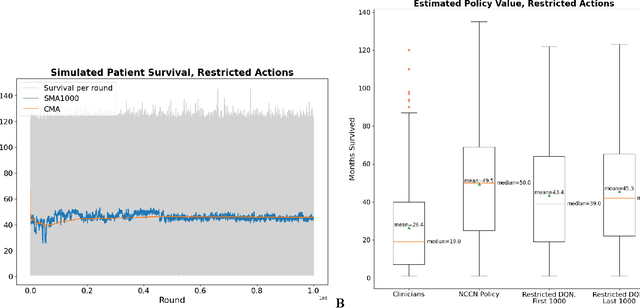
Abstract:The prognosis for patients with epithelial ovarian cancer remains dismal despite improvements in survival for other cancers. Treatment involves multiple lines of chemotherapy and becomes increasingly heterogeneous after first-line therapy. Reinforcement learning with real-world outcomes data has the potential to identify novel treatment strategies to improve overall survival. We design a reinforcement learning environment to model epithelial ovarian cancer treatment trajectories and use model free reinforcement learning to investigate therapeutic regimens for simulated patients.
Learn like a Pathologist: Curriculum Learning by Annotator Agreement for Histopathology Image Classification
Sep 29, 2020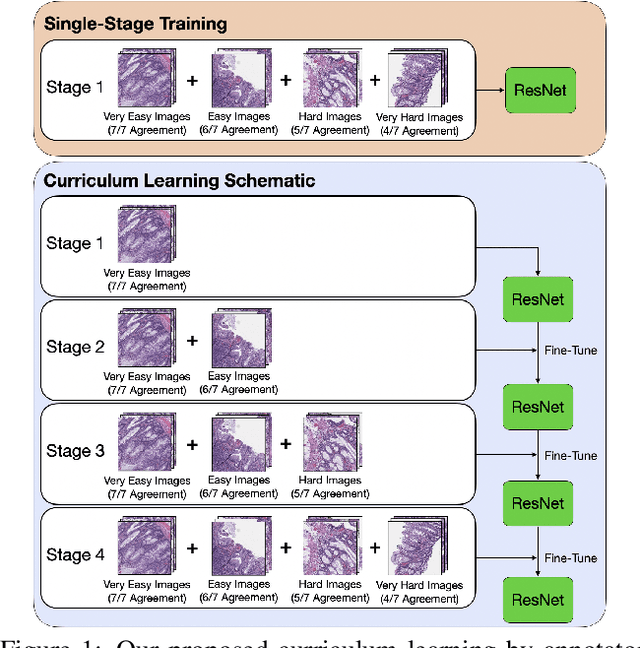
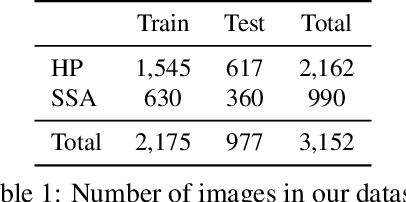
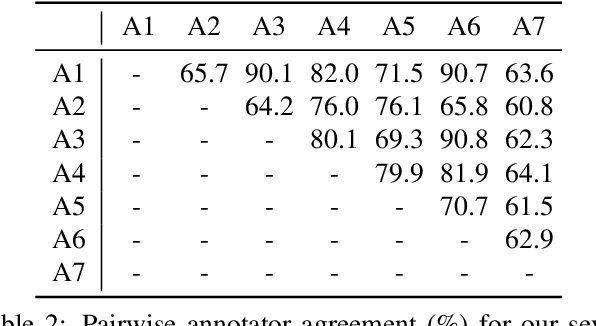
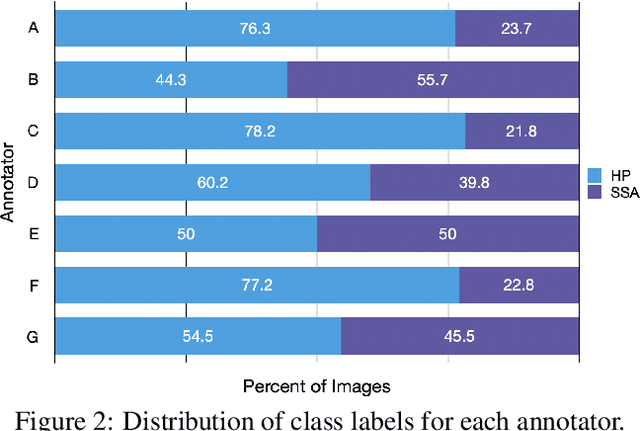
Abstract:Applying curriculum learning requires both a range of difficulty in data and a method for determining the difficulty of examples. In many tasks, however, satisfying these requirements can be a formidable challenge. In this paper, we contend that histopathology image classification is a compelling use case for curriculum learning. Based on the nature of histopathology images, a range of difficulty inherently exists among examples, and, since medical datasets are often labeled by multiple annotators, annotator agreement can be used as a natural proxy for the difficulty of a given example. Hence, we propose a simple curriculum learning method that trains on progressively-harder images as determined by annotator agreement. We evaluate our hypothesis on the challenging and clinically-important task of colorectal polyp classification. Whereas vanilla training achieves an AUC of 83.7% for this task, a model trained with our proposed curriculum learning approach achieves an AUC of 88.2%, an improvement of 4.5%. Our work aims to inspire researchers to think more creatively and rigorously when choosing contexts for applying curriculum learning.
Difficulty Translation in Histopathology Images
Apr 27, 2020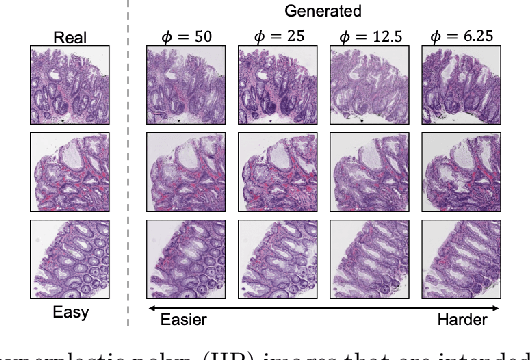
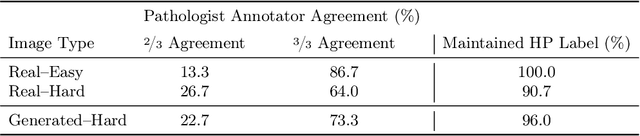
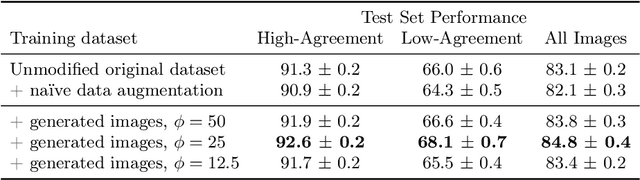
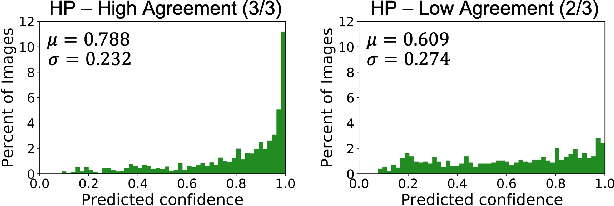
Abstract:The unique nature of histopathology images opens the door to domain-specific formulations of image translation models. We propose a difficulty translation model that modifies colorectal histopathology images to become more challenging to classify. Our model comprises a scorer, which provides an output confidence to measure the difficulty of images, and an image translator, which learns to translate images from easy-to-classify to hard-to-classify using a training set defined by the scorer. We present three findings. First, generated images were indeed harder to classify for both human pathologists and machine learning classifiers than their corresponding source images. Second, image classifiers trained with generated images as augmented data performed better on both easy and hard images from an independent test set. Finally, human annotator agreement and our model's measure of difficulty correlated strongly, implying that for future work requiring human annotator agreement, the confidence score of a machine learning classifier could be used instead as a proxy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge