Afshin Bozorgpour
CENet: Context Enhancement Network for Medical Image Segmentation
May 23, 2025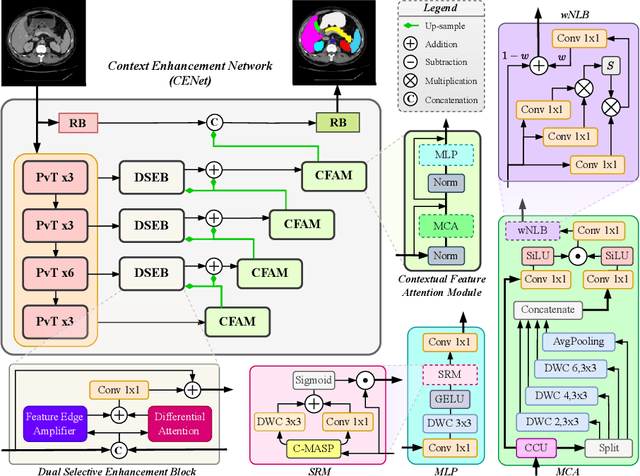
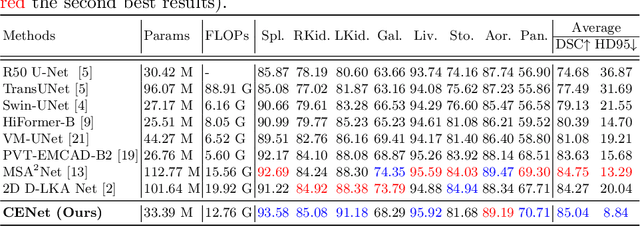
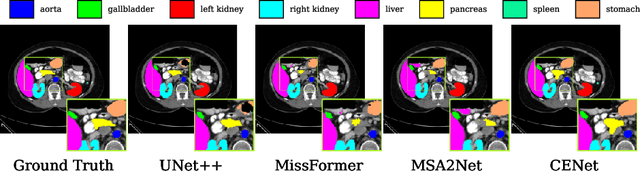
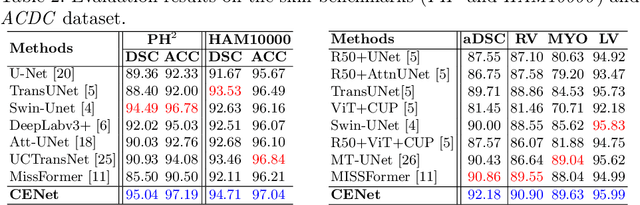
Abstract:Medical image segmentation, particularly in multi-domain scenarios, requires precise preservation of anatomical structures across diverse representations. While deep learning has advanced this field, existing models often struggle with accurate boundary representation, variability in organ morphology, and information loss during downsampling, limiting their accuracy and robustness. To address these challenges, we propose the Context Enhancement Network (CENet), a novel segmentation framework featuring two key innovations. First, the Dual Selective Enhancement Block (DSEB) integrated into skip connections enhances boundary details and improves the detection of smaller organs in a context-aware manner. Second, the Context Feature Attention Module (CFAM) in the decoder employs a multi-scale design to maintain spatial integrity, reduce feature redundancy, and mitigate overly enhanced representations. Extensive evaluations on both radiology and dermoscopic datasets demonstrate that CENet outperforms state-of-the-art (SOTA) methods in multi-organ segmentation and boundary detail preservation, offering a robust and accurate solution for complex medical image analysis tasks. The code is publicly available at https://github.com/xmindflow/cenet.
Attention-based Generative Latent Replay: A Continual Learning Approach for WSI Analysis
May 13, 2025



Abstract:Whole slide image (WSI) classification has emerged as a powerful tool in computational pathology, but remains constrained by domain shifts, e.g., due to different organs, diseases, or institution-specific variations. To address this challenge, we propose an Attention-based Generative Latent Replay Continual Learning framework (AGLR-CL), in a multiple instance learning (MIL) setup for domain incremental WSI classification. Our method employs Gaussian Mixture Models (GMMs) to synthesize WSI representations and patch count distributions, preserving knowledge of past domains without explicitly storing original data. A novel attention-based filtering step focuses on the most salient patch embeddings, ensuring high-quality synthetic samples. This privacy-aware strategy obviates the need for replay buffers and outperforms other buffer-free counterparts while matching the performance of buffer-based solutions. We validate AGLR-CL on clinically relevant biomarker detection and molecular status prediction across multiple public datasets with diverse centers, organs, and patient cohorts. Experimental results confirm its ability to retain prior knowledge and adapt to new domains, offering an effective, privacy-preserving avenue for domain incremental continual learning in WSI classification.
Domain-incremental White Blood Cell Classification with Privacy-aware Continual Learning
Mar 25, 2025Abstract:White blood cell (WBC) classification plays a vital role in hematology for diagnosing various medical conditions. However, it faces significant challenges due to domain shifts caused by variations in sample sources (e.g., blood or bone marrow) and differing imaging conditions across hospitals. Traditional deep learning models often suffer from catastrophic forgetting in such dynamic environments, while foundation models, though generally robust, experience performance degradation when the distribution of inference data differs from that of the training data. To address these challenges, we propose a generative replay-based Continual Learning (CL) strategy designed to prevent forgetting in foundation models for WBC classification. Our method employs lightweight generators to mimic past data with a synthetic latent representation to enable privacy-preserving replay. To showcase the effectiveness, we carry out extensive experiments with a total of four datasets with different task ordering and four backbone models including ResNet50, RetCCL, CTransPath, and UNI. Experimental results demonstrate that conventional fine-tuning methods degrade performance on previously learned tasks and struggle with domain shifts. In contrast, our continual learning strategy effectively mitigates catastrophic forgetting, preserving model performance across varying domains. This work presents a practical solution for maintaining reliable WBC classification in real-world clinical settings, where data distributions frequently evolve.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
MSA$^2$Net: Multi-scale Adaptive Attention-guided Network for Medical Image Segmentation
Aug 03, 2024Abstract:Medical image segmentation involves identifying and separating object instances in a medical image to delineate various tissues and structures, a task complicated by the significant variations in size, shape, and density of these features. Convolutional neural networks (CNNs) have traditionally been used for this task but have limitations in capturing long-range dependencies. Transformers, equipped with self-attention mechanisms, aim to address this problem. However, in medical image segmentation it is beneficial to merge both local and global features to effectively integrate feature maps across various scales, capturing both detailed features and broader semantic elements for dealing with variations in structures. In this paper, we introduce MSA$^2$Net, a new deep segmentation framework featuring an expedient design of skip-connections. These connections facilitate feature fusion by dynamically weighting and combining coarse-grained encoder features with fine-grained decoder feature maps. Specifically, we propose a Multi-Scale Adaptive Spatial Attention Gate (MASAG), which dynamically adjusts the receptive field (Local and Global contextual information) to ensure that spatially relevant features are selectively highlighted while minimizing background distractions. Extensive evaluations involving dermatology, and radiological datasets demonstrate that our MSA$^2$Net outperforms state-of-the-art (SOTA) works or matches their performance. The source code is publicly available at https://github.com/xmindflow/MSA-2Net.
MSA2Net: Multi-scale Adaptive Attention-guided Network for Medical Image Segmentation
Jul 31, 2024Abstract:Medical image segmentation involves identifying and separating object instances in a medical image to delineate various tissues and structures, a task complicated by the significant variations in size, shape, and density of these features. Convolutional neural networks (CNNs) have traditionally been used for this task but have limitations in capturing long-range dependencies. Transformers, equipped with self-attention mechanisms, aim to address this problem. However, in medical image segmentation it is beneficial to merge both local and global features to effectively integrate feature maps across various scales, capturing both detailed features and broader semantic elements for dealing with variations in structures. In this paper, we introduce MSA2Net, a new deep segmentation framework featuring an expedient design of skip-connections. These connections facilitate feature fusion by dynamically weighting and combining coarse-grained encoder features with fine-grained decoder feature maps. Specifically, we propose a Multi-Scale Adaptive Spatial Attention Gate (MASAG), which dynamically adjusts the receptive field (Local and Global contextual information) to ensure that spatially relevant features are selectively highlighted while minimizing background distractions. Extensive evaluations involving dermatology, and radiological datasets demonstrate that our MSA2Net outperforms state-of-the-art (SOTA) works or matches their performance. The source code is publicly available at https://github.com/xmindflow/MSA-2Net.
Physics-Inspired Generative Models in Medical Imaging: A Review
Jul 15, 2024


Abstract:Physics-inspired generative models, in particular diffusion and Poisson flow models, enhance Bayesian methods and promise great utilities in medical imaging. This review examines the transformative role of such generative methods. First, a variety of physics-inspired generative models, including Denoising Diffusion Probabilistic Models (DDPM), Score-based Diffusion Models, and Poisson Flow Generative Models (PFGM and PFGM++), are revisited, with an emphasis on their accuracy, robustness as well as acceleration. Then, major applications of physics-inspired generative models in medical imaging are presented, comprising image reconstruction, image generation, and image analysis. Finally, future research directions are brainstormed, including unification of physics-inspired generative models, integration with vision-language models (VLMs),and potential novel applications of generative models. Since the development of generative methods has been rapid, this review will hopefully give peers and learners a timely snapshot of this new family of physics-driven generative models and help capitalize their enormous potential for medical imaging.
Computation-Efficient Era: A Comprehensive Survey of State Space Models in Medical Image Analysis
Jun 05, 2024



Abstract:Sequence modeling plays a vital role across various domains, with recurrent neural networks being historically the predominant method of performing these tasks. However, the emergence of transformers has altered this paradigm due to their superior performance. Built upon these advances, transformers have conjoined CNNs as two leading foundational models for learning visual representations. However, transformers are hindered by the $\mathcal{O}(N^2)$ complexity of their attention mechanisms, while CNNs lack global receptive fields and dynamic weight allocation. State Space Models (SSMs), specifically the \textit{\textbf{Mamba}} model with selection mechanisms and hardware-aware architecture, have garnered immense interest lately in sequential modeling and visual representation learning, challenging the dominance of transformers by providing infinite context lengths and offering substantial efficiency maintaining linear complexity in the input sequence. Capitalizing on the advances in computer vision, medical imaging has heralded a new epoch with Mamba models. Intending to help researchers navigate the surge, this survey seeks to offer an encyclopedic review of Mamba models in medical imaging. Specifically, we start with a comprehensive theoretical review forming the basis of SSMs, including Mamba architecture and its alternatives for sequence modeling paradigms in this context. Next, we offer a structured classification of Mamba models in the medical field and introduce a diverse categorization scheme based on their application, imaging modalities, and targeted organs. Finally, we summarize key challenges, discuss different future research directions of the SSMs in the medical domain, and propose several directions to fulfill the demands of this field. In addition, we have compiled the studies discussed in this paper along with their open-source implementations on our GitHub repository.
LHU-Net: A Light Hybrid U-Net for Cost-Efficient, High-Performance Volumetric Medical Image Segmentation
Apr 07, 2024Abstract:As a result of the rise of Transformer architectures in medical image analysis, specifically in the domain of medical image segmentation, a multitude of hybrid models have been created that merge the advantages of Convolutional Neural Networks (CNNs) and Transformers. These hybrid models have achieved notable success by significantly improving segmentation accuracy. Yet, this progress often comes at the cost of increased model complexity, both in terms of parameters and computational demand. Moreover, many of these models fail to consider the crucial interplay between spatial and channel features, which could further refine and improve segmentation outcomes. To address this, we introduce LHU-Net, a Light Hybrid U-Net architecture optimized for volumetric medical image segmentation. LHU-Net is meticulously designed to prioritize spatial feature analysis in its initial layers before shifting focus to channel-based features in its deeper layers, ensuring a comprehensive feature extraction process. Rigorous evaluation across five benchmark datasets - Synapse, LA, Pancreas, ACDC, and BRaTS 2018 - underscores LHU-Net's superior performance, showcasing its dual capacity for efficiency and accuracy. Notably, LHU-Net sets new performance benchmarks, such as attaining a Dice score of 92.66 on the ACDC dataset, while simultaneously reducing parameters by 85% and quartering the computational load compared to existing state-of-the-art models. Achieved without any reliance on pre-training, additional data, or model ensemble, LHU-Net's effectiveness is further evidenced by its state-of-the-art performance across all evaluated datasets, utilizing fewer than 11 million parameters. This achievement highlights that balancing computational efficiency with high accuracy in medical image segmentation is feasible. Our implementation of LHU-Net is freely accessible to the research community on GitHub.
Enhancing Efficiency in Vision Transformer Networks: Design Techniques and Insights
Mar 28, 2024



Abstract:Intrigued by the inherent ability of the human visual system to identify salient regions in complex scenes, attention mechanisms have been seamlessly integrated into various Computer Vision (CV) tasks. Building upon this paradigm, Vision Transformer (ViT) networks exploit attention mechanisms for improved efficiency. This review navigates the landscape of redesigned attention mechanisms within ViTs, aiming to enhance their performance. This paper provides a comprehensive exploration of techniques and insights for designing attention mechanisms, systematically reviewing recent literature in the field of CV. This survey begins with an introduction to the theoretical foundations and fundamental concepts underlying attention mechanisms. We then present a systematic taxonomy of various attention mechanisms within ViTs, employing redesigned approaches. A multi-perspective categorization is proposed based on their application, objectives, and the type of attention applied. The analysis includes an exploration of the novelty, strengths, weaknesses, and an in-depth evaluation of the different proposed strategies. This culminates in the development of taxonomies that highlight key properties and contributions. Finally, we gather the reviewed studies along with their available open-source implementations at our \href{https://github.com/mindflow-institue/Awesome-Attention-Mechanism-in-Medical-Imaging}{GitHub}\footnote{\url{https://github.com/xmindflow/Awesome-Attention-Mechanism-in-Medical-Imaging}}. We aim to regularly update it with the most recent relevant papers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge