Ilker Hacihaliloglu
CENet: Context Enhancement Network for Medical Image Segmentation
May 23, 2025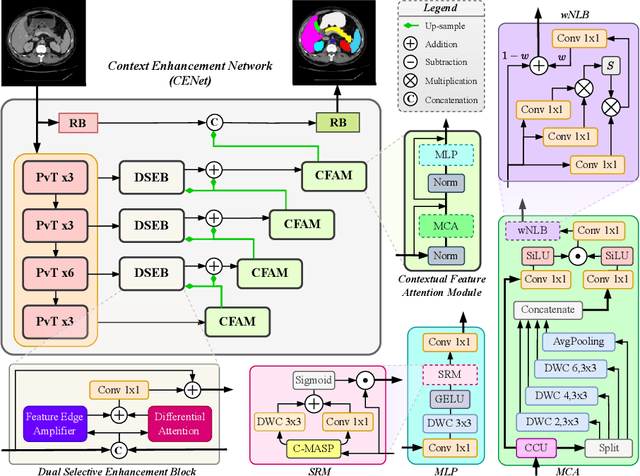
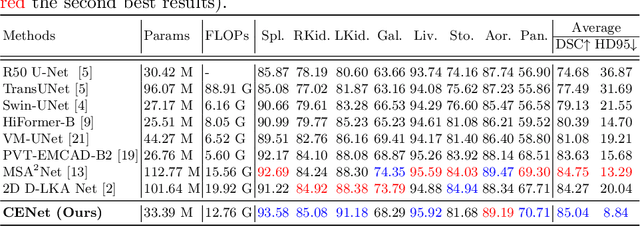
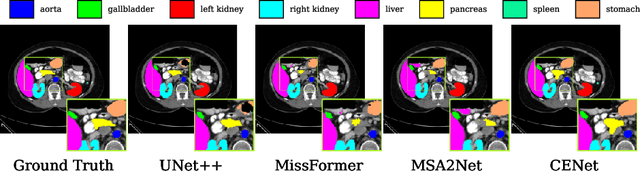
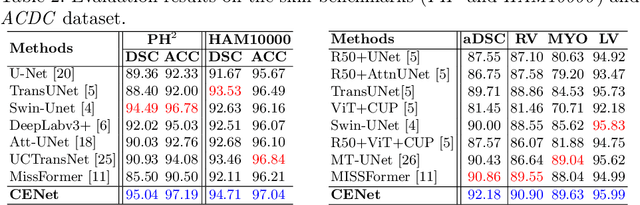
Abstract:Medical image segmentation, particularly in multi-domain scenarios, requires precise preservation of anatomical structures across diverse representations. While deep learning has advanced this field, existing models often struggle with accurate boundary representation, variability in organ morphology, and information loss during downsampling, limiting their accuracy and robustness. To address these challenges, we propose the Context Enhancement Network (CENet), a novel segmentation framework featuring two key innovations. First, the Dual Selective Enhancement Block (DSEB) integrated into skip connections enhances boundary details and improves the detection of smaller organs in a context-aware manner. Second, the Context Feature Attention Module (CFAM) in the decoder employs a multi-scale design to maintain spatial integrity, reduce feature redundancy, and mitigate overly enhanced representations. Extensive evaluations on both radiology and dermoscopic datasets demonstrate that CENet outperforms state-of-the-art (SOTA) methods in multi-organ segmentation and boundary detail preservation, offering a robust and accurate solution for complex medical image analysis tasks. The code is publicly available at https://github.com/xmindflow/cenet.
AllMetrics: A Unified Python Library for Standardized Metric Evaluation and Robust Data Validation in Machine Learning
May 21, 2025Abstract:Machine learning (ML) models rely heavily on consistent and accurate performance metrics to evaluate and compare their effectiveness. However, existing libraries often suffer from fragmentation, inconsistent implementations, and insufficient data validation protocols, leading to unreliable results. Existing libraries have often been developed independently and without adherence to a unified standard, particularly concerning the specific tasks they aim to support. As a result, each library tends to adopt its conventions for metric computation, input/output formatting, error handling, and data validation protocols. This lack of standardization leads to both implementation differences (ID) and reporting differences (RD), making it difficult to compare results across frameworks or ensure reliable evaluations. To address these issues, we introduce AllMetrics, an open-source unified Python library designed to standardize metric evaluation across diverse ML tasks, including regression, classification, clustering, segmentation, and image-to-image translation. The library implements class-specific reporting for multi-class tasks through configurable parameters to cover all use cases, while incorporating task-specific parameters to resolve metric computation discrepancies across implementations. Various datasets from domains like healthcare, finance, and real estate were applied to our library and compared with Python, Matlab, and R components to identify which yield similar results. AllMetrics combines a modular Application Programming Interface (API) with robust input validation mechanisms to ensure reproducibility and reliability in model evaluation. This paper presents the design principles, architectural components, and empirical analyses demonstrating the ability to mitigate evaluation errors and to enhance the trustworthiness of ML workflows.
Pathobiological Dictionary Defining Pathomics and Texture Features: Addressing Understandable AI Issues in Personalized Liver Cancer; Dictionary Version LCP1.0
May 20, 2025Abstract:Artificial intelligence (AI) holds strong potential for medical diagnostics, yet its clinical adoption is limited by a lack of interpretability and generalizability. This study introduces the Pathobiological Dictionary for Liver Cancer (LCP1.0), a practical framework designed to translate complex Pathomics and Radiomics Features (PF and RF) into clinically meaningful insights aligned with existing diagnostic workflows. QuPath and PyRadiomics, standardized according to IBSI guidelines, were used to extract 333 imaging features from hepatocellular carcinoma (HCC) tissue samples, including 240 PF-based-cell detection/intensity, 74 RF-based texture, and 19 RF-based first-order features. Expert-defined ROIs from the public dataset excluded artifact-prone areas, and features were aggregated at the case level. Their relevance to the WHO grading system was assessed using multiple classifiers linked with feature selectors. The resulting dictionary was validated by 8 experts in oncology and pathology. In collaboration with 10 domain experts, we developed a Pathobiological dictionary of imaging features such as PFs and RF. In our study, the Variable Threshold feature selection algorithm combined with the SVM model achieved the highest accuracy (0.80, P-value less than 0.05), selecting 20 key features, primarily clinical and pathomics traits such as Centroid, Cell Nucleus, and Cytoplasmic characteristics. These features, particularly nuclear and cytoplasmic, were strongly associated with tumor grading and prognosis, reflecting atypia indicators like pleomorphism, hyperchromasia, and cellular orientation.The LCP1.0 provides a clinically validated bridge between AI outputs and expert interpretation, enhancing model transparency and usability. Aligning AI-derived features with clinical semantics supports the development of interpretable, trustworthy diagnostic tools for liver cancer pathology.
Impact of Data Patterns on Biotype identification Using Machine Learning
Mar 15, 2025



Abstract:Background: Patient stratification in brain disorders remains a significant challenge, despite advances in machine learning and multimodal neuroimaging. Automated machine learning algorithms have been widely applied for identifying patient subtypes (biotypes), but results have been inconsistent across studies. These inconsistencies are often attributed to algorithmic limitations, yet an overlooked factor may be the statistical properties of the input data. This study investigates the contribution of data patterns on algorithm performance by leveraging synthetic brain morphometry data as an exemplar. Methods: Four widely used algorithms-SuStaIn, HYDRA, SmileGAN, and SurrealGAN were evaluated using multiple synthetic pseudo-patient datasets designed to include varying numbers and sizes of clusters and degrees of complexity of morphometric changes. Ground truth, representing predefined clusters, allowed for the evaluation of performance accuracy across algorithms and datasets. Results: SuStaIn failed to process datasets with more than 17 variables, highlighting computational inefficiencies. HYDRA was able to perform individual-level classification in multiple datasets with no clear pattern explaining failures. SmileGAN and SurrealGAN outperformed other algorithms in identifying variable-based disease patterns, but these patterns were not able to provide individual-level classification. Conclusions: Dataset characteristics significantly influence algorithm performance, often more than algorithmic design. The findings emphasize the need for rigorous validation using synthetic data before real-world application and highlight the limitations of current clustering approaches in capturing the heterogeneity of brain disorders. These insights extend beyond neuroimaging and have implications for machine learning applications in biomedical research.
Mono2D: A Trainable Monogenic Layer for Robust Knee Cartilage Segmentation on Out-of-Distribution 2D Ultrasound Data
Mar 12, 2025



Abstract:Automated knee cartilage segmentation using point-of-care ultrasound devices and deep-learning networks has the potential to enhance the management of knee osteoarthritis. However, segmentation algorithms often struggle with domain shifts caused by variations in ultrasound devices and acquisition parameters, limiting their generalizability. In this paper, we propose Mono2D, a monogenic layer that extracts multi-scale, contrast- and intensity-invariant local phase features using trainable bandpass quadrature filters. This layer mitigates domain shifts, improving generalization to out-of-distribution domains. Mono2D is integrated before the first layer of a segmentation network, and its parameters jointly trained alongside the network's parameters. We evaluated Mono2D on a multi-domain 2D ultrasound knee cartilage dataset for single-source domain generalization (SSDG). Our results demonstrate that Mono2D outperforms other SSDG methods in terms of Dice score and mean average surface distance. To further assess its generalizability, we evaluate Mono2D on a multi-site prostate MRI dataset, where it continues to outperform other SSDG methods, highlighting its potential to improve domain generalization in medical imaging. Nevertheless, further evaluation on diverse datasets is still necessary to assess its clinical utility.
Influence of High-Performance Image-to-Image Translation Networks on Clinical Visual Assessment and Outcome Prediction: Utilizing Ultrasound to MRI Translation in Prostate Cancer
Jan 30, 2025



Abstract:Purpose: This study examines the core traits of image-to-image translation (I2I) networks, focusing on their effectiveness and adaptability in everyday clinical settings. Methods: We have analyzed data from 794 patients diagnosed with prostate cancer (PCa), using ten prominent 2D/3D I2I networks to convert ultrasound (US) images into MRI scans. We also introduced a new analysis of Radiomic features (RF) via the Spearman correlation coefficient to explore whether networks with high performance (SSIM>85%) could detect subtle RFs. Our study further examined synthetic images by 7 invited physicians. As a final evaluation study, we have investigated the improvement that are achieved using the synthetic MRI data on two traditional machine learning and one deep learning method. Results: In quantitative assessment, 2D-Pix2Pix network substantially outperformed the other 7 networks, with an average SSIM~0.855. The RF analysis revealed that 76 out of 186 RFs were identified using the 2D-Pix2Pix algorithm alone, although half of the RFs were lost during the translation process. A detailed qualitative review by 7 medical doctors noted a deficiency in low-level feature recognition in I2I tasks. Furthermore, the study found that synthesized image-based classification outperformed US image-based classification with an average accuracy and AUC~0.93. Conclusion: This study showed that while 2D-Pix2Pix outperformed cutting-edge networks in low-level feature discovery and overall error and similarity metrics, it still requires improvement in low-level feature performance, as highlighted by Group 3. Further, the study found using synthetic image-based classification outperformed original US image-based methods.
Biological and Radiological Dictionary of Radiomics Features: Addressing Understandable AI Issues in Personalized Prostate Cancer; Dictionary version PM1.0
Dec 14, 2024



Abstract:This study investigates the connection between visual semantic features in PI-RADS and associated risk factors, moving beyond abnormal imaging findings by creating a standardized dictionary of biological/radiological radiomics features (RFs). Using multiparametric prostate MRI sequences (T2-weighted imaging [T2WI], diffusion-weighted imaging [DWI], and apparent diffusion coefficient [ADC]), six interpretable and seven complex classifiers, combined with nine feature selection algorithms (FSAs), were applied to segmented lesions to predict UCLA scores. Combining T2WI, DWI, and ADC with FSAs such as ANOVA F-test, Correlation Coefficient, and Fisher Score, and utilizing logistic regression, identified key features: the 90th percentile from T2WI (hypo-intensity linked to cancer risk), variance from T2WI (lesion heterogeneity), shape metrics like Least Axis Length and Surface Area to Volume ratio from ADC (lesion compactness), and Run Entropy from ADC (texture consistency). This approach achieved an average accuracy of 0.78, outperforming single-sequence methods (p < 0.05). The developed dictionary provides a common language, fostering collaboration between clinical professionals and AI developers to enable trustworthy, interpretable AI for reliable clinical decisions.
Prompt2Perturb (P2P): Text-Guided Diffusion-Based Adversarial Attacks on Breast Ultrasound Images
Dec 13, 2024Abstract:Deep neural networks (DNNs) offer significant promise for improving breast cancer diagnosis in medical imaging. However, these models are highly susceptible to adversarial attacks--small, imperceptible changes that can mislead classifiers--raising critical concerns about their reliability and security. Traditional attacks rely on fixed-norm perturbations, misaligning with human perception. In contrast, diffusion-based attacks require pre-trained models, demanding substantial data when these models are unavailable, limiting practical use in data-scarce scenarios. In medical imaging, however, this is often unfeasible due to the limited availability of datasets. Building on recent advancements in learnable prompts, we propose Prompt2Perturb (P2P), a novel language-guided attack method capable of generating meaningful attack examples driven by text instructions. During the prompt learning phase, our approach leverages learnable prompts within the text encoder to create subtle, yet impactful, perturbations that remain imperceptible while guiding the model towards targeted outcomes. In contrast to current prompt learning-based approaches, our P2P stands out by directly updating text embeddings, avoiding the need for retraining diffusion models. Further, we leverage the finding that optimizing only the early reverse diffusion steps boosts efficiency while ensuring that the generated adversarial examples incorporate subtle noise, thus preserving ultrasound image quality without introducing noticeable artifacts. We show that our method outperforms state-of-the-art attack techniques across three breast ultrasound datasets in FID and LPIPS. Moreover, the generated images are both more natural in appearance and more effective compared to existing adversarial attacks. Our code will be publicly available https://github.com/yasamin-med/P2P.
Machine Learning Evaluation Metric Discrepancies across Programming Languages and Their Components: Need for Standardization
Nov 18, 2024



Abstract:This study evaluates metrics for tasks such as classification, regression, clustering, correlation analysis, statistical tests, segmentation, and image-to-image (I2I) translation. Metrics were compared across Python libraries, R packages, and Matlab functions to assess their consistency and highlight discrepancies. The findings underscore the need for a unified roadmap to standardize metrics, ensuring reliable and reproducible ML evaluations across platforms. This study examined a wide range of evaluation metrics across various tasks and found only some to be consistent across platforms, such as (i) Accuracy, Balanced Accuracy, Cohens Kappa, F-beta Score, MCC, Geometric Mean, AUC, and Log Loss in binary classification; (ii) Accuracy, Cohens Kappa, and F-beta Score in multi-class classification; (iii) MAE, MSE, RMSE, MAPE, Explained Variance, Median AE, MSLE, and Huber in regression; (iv) Davies-Bouldin Index and Calinski-Harabasz Index in clustering; (v) Pearson, Spearman, Kendall's Tau, Mutual Information, Distance Correlation, Percbend, Shepherd, and Partial Correlation in correlation analysis; (vi) Paired t-test, Chi-Square Test, ANOVA, Kruskal-Wallis Test, Shapiro-Wilk Test, Welchs t-test, and Bartlett's test in statistical tests; (vii) Accuracy, Precision, and Recall in 2D segmentation; (viii) Accuracy in 3D segmentation; (ix) MAE, MSE, RMSE, and R-Squared in 2D-I2I translation; and (x) MAE, MSE, and RMSE in 3D-I2I translation. Given observation of discrepancies in a number of metrics (e.g. precision, recall and F1 score in binary classification, WCSS in clustering, multiple statistical tests, and IoU in segmentation, amongst multiple metrics), this study concludes that ML evaluation metrics require standardization and recommends that future research use consistent metrics for different tasks to effectively compare ML techniques and solutions.
Single-Layer Learnable Activation for Implicit Neural Representation (SL$^{2}$A-INR)
Sep 18, 2024



Abstract:Implicit Neural Representation (INR), leveraging a neural network to transform coordinate input into corresponding attributes, has recently driven significant advances in several vision-related domains. However, the performance of INR is heavily influenced by the choice of the nonlinear activation function used in its multilayer perceptron (MLP) architecture. Multiple nonlinearities have been investigated; yet, current INRs face limitations in capturing high-frequency components, diverse signal types, and handling inverse problems. We have identified that these problems can be greatly alleviated by introducing a paradigm shift in INRs. We find that an architecture with learnable activations in initial layers can represent fine details in the underlying signals. Specifically, we propose SL$^{2}$A-INR, a hybrid network for INR with a single-layer learnable activation function, prompting the effectiveness of traditional ReLU-based MLPs. Our method performs superior across diverse tasks, including image representation, 3D shape reconstructions, inpainting, single image super-resolution, CT reconstruction, and novel view synthesis. Through comprehensive experiments, SL$^{2}$A-INR sets new benchmarks in accuracy, quality, and convergence rates for INR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge