Sanaz Karimijafarbigloo
Computation-Efficient Era: A Comprehensive Survey of State Space Models in Medical Image Analysis
Jun 05, 2024



Abstract:Sequence modeling plays a vital role across various domains, with recurrent neural networks being historically the predominant method of performing these tasks. However, the emergence of transformers has altered this paradigm due to their superior performance. Built upon these advances, transformers have conjoined CNNs as two leading foundational models for learning visual representations. However, transformers are hindered by the $\mathcal{O}(N^2)$ complexity of their attention mechanisms, while CNNs lack global receptive fields and dynamic weight allocation. State Space Models (SSMs), specifically the \textit{\textbf{Mamba}} model with selection mechanisms and hardware-aware architecture, have garnered immense interest lately in sequential modeling and visual representation learning, challenging the dominance of transformers by providing infinite context lengths and offering substantial efficiency maintaining linear complexity in the input sequence. Capitalizing on the advances in computer vision, medical imaging has heralded a new epoch with Mamba models. Intending to help researchers navigate the surge, this survey seeks to offer an encyclopedic review of Mamba models in medical imaging. Specifically, we start with a comprehensive theoretical review forming the basis of SSMs, including Mamba architecture and its alternatives for sequence modeling paradigms in this context. Next, we offer a structured classification of Mamba models in the medical field and introduce a diverse categorization scheme based on their application, imaging modalities, and targeted organs. Finally, we summarize key challenges, discuss different future research directions of the SSMs in the medical domain, and propose several directions to fulfill the demands of this field. In addition, we have compiled the studies discussed in this paper along with their open-source implementations on our GitHub repository.
Loss Functions in the Era of Semantic Segmentation: A Survey and Outlook
Dec 08, 2023



Abstract:Semantic image segmentation, the process of classifying each pixel in an image into a particular class, plays an important role in many visual understanding systems. As the predominant criterion for evaluating the performance of statistical models, loss functions are crucial for shaping the development of deep learning-based segmentation algorithms and improving their overall performance. To aid researchers in identifying the optimal loss function for their particular application, this survey provides a comprehensive and unified review of $25$ loss functions utilized in image segmentation. We provide a novel taxonomy and thorough review of how these loss functions are customized and leveraged in image segmentation, with a systematic categorization emphasizing their significant features and applications. Furthermore, to evaluate the efficacy of these methods in real-world scenarios, we propose unbiased evaluations of some distinct and renowned loss functions on established medical and natural image datasets. We conclude this review by identifying current challenges and unveiling future research opportunities. Finally, we have compiled the reviewed studies that have open-source implementations on our GitHub page.
FuseNet: Self-Supervised Dual-Path Network for Medical Image Segmentation
Nov 22, 2023Abstract:Semantic segmentation, a crucial task in computer vision, often relies on labor-intensive and costly annotated datasets for training. In response to this challenge, we introduce FuseNet, a dual-stream framework for self-supervised semantic segmentation that eliminates the need for manual annotation. FuseNet leverages the shared semantic dependencies between the original and augmented images to create a clustering space, effectively assigning pixels to semantically related clusters, and ultimately generating the segmentation map. Additionally, FuseNet incorporates a cross-modal fusion technique that extends the principles of CLIP by replacing textual data with augmented images. This approach enables the model to learn complex visual representations, enhancing robustness against variations similar to CLIP's text invariance. To further improve edge alignment and spatial consistency between neighboring pixels, we introduce an edge refinement loss. This loss function considers edge information to enhance spatial coherence, facilitating the grouping of nearby pixels with similar visual features. Extensive experiments on skin lesion and lung segmentation datasets demonstrate the effectiveness of our method. \href{https://github.com/xmindflow/FuseNet}{Codebase.}
Leveraging Unlabeled Data for 3D Medical Image Segmentation through Self-Supervised Contrastive Learning
Nov 21, 2023Abstract:Current 3D semi-supervised segmentation methods face significant challenges such as limited consideration of contextual information and the inability to generate reliable pseudo-labels for effective unsupervised data use. To address these challenges, we introduce two distinct subnetworks designed to explore and exploit the discrepancies between them, ultimately correcting the erroneous prediction results. More specifically, we identify regions of inconsistent predictions and initiate a targeted verification training process. This procedure strategically fine-tunes and harmonizes the predictions of the subnetworks, leading to enhanced utilization of contextual information. Furthermore, to adaptively fine-tune the network's representational capacity and reduce prediction uncertainty, we employ a self-supervised contrastive learning paradigm. For this, we use the network's confidence to distinguish between reliable and unreliable predictions. The model is then trained to effectively minimize unreliable predictions. Our experimental results for organ segmentation, obtained from clinical MRI and CT scans, demonstrate the effectiveness of our approach when compared to state-of-the-art methods. The codebase is accessible on \href{https://github.com/xmindflow/SSL-contrastive}{GitHub}.
Self-supervised Semantic Segmentation: Consistency over Transformation
Aug 31, 2023Abstract:Accurate medical image segmentation is of utmost importance for enabling automated clinical decision procedures. However, prevailing supervised deep learning approaches for medical image segmentation encounter significant challenges due to their heavy dependence on extensive labeled training data. To tackle this issue, we propose a novel self-supervised algorithm, \textbf{S$^3$-Net}, which integrates a robust framework based on the proposed Inception Large Kernel Attention (I-LKA) modules. This architectural enhancement makes it possible to comprehensively capture contextual information while preserving local intricacies, thereby enabling precise semantic segmentation. Furthermore, considering that lesions in medical images often exhibit deformations, we leverage deformable convolution as an integral component to effectively capture and delineate lesion deformations for superior object boundary definition. Additionally, our self-supervised strategy emphasizes the acquisition of invariance to affine transformations, which is commonly encountered in medical scenarios. This emphasis on robustness with respect to geometric distortions significantly enhances the model's ability to accurately model and handle such distortions. To enforce spatial consistency and promote the grouping of spatially connected image pixels with similar feature representations, we introduce a spatial consistency loss term. This aids the network in effectively capturing the relationships among neighboring pixels and enhancing the overall segmentation quality. The S$^3$-Net approach iteratively learns pixel-level feature representations for image content clustering in an end-to-end manner. Our experimental results on skin lesion and lung organ segmentation tasks show the superior performance of our method compared to the SOTA approaches. https://github.com/mindflow-institue/SSCT
Self-supervised Few-shot Learning for Semantic Segmentation: An Annotation-free Approach
Jul 26, 2023



Abstract:Few-shot semantic segmentation (FSS) offers immense potential in the field of medical image analysis, enabling accurate object segmentation with limited training data. However, existing FSS techniques heavily rely on annotated semantic classes, rendering them unsuitable for medical images due to the scarcity of annotations. To address this challenge, multiple contributions are proposed: First, inspired by spectral decomposition methods, the problem of image decomposition is reframed as a graph partitioning task. The eigenvectors of the Laplacian matrix, derived from the feature affinity matrix of self-supervised networks, are analyzed to estimate the distribution of the objects of interest from the support images. Secondly, we propose a novel self-supervised FSS framework that does not rely on any annotation. Instead, it adaptively estimates the query mask by leveraging the eigenvectors obtained from the support images. This approach eliminates the need for manual annotation, making it particularly suitable for medical images with limited annotated data. Thirdly, to further enhance the decoding of the query image based on the information provided by the support image, we introduce a multi-scale large kernel attention module. By selectively emphasizing relevant features and details, this module improves the segmentation process and contributes to better object delineation. Evaluations on both natural and medical image datasets demonstrate the efficiency and effectiveness of our method. Moreover, the proposed approach is characterized by its generality and model-agnostic nature, allowing for seamless integration with various deep architectures. The code is publicly available at \href{https://github.com/mindflow-institue/annotation_free_fewshot}{\textcolor{magenta}{GitHub}}.
Medical Image Segmentation Review: The success of U-Net
Nov 27, 2022



Abstract:Automatic medical image segmentation is a crucial topic in the medical domain and successively a critical counterpart in the computer-aided diagnosis paradigm. U-Net is the most widespread image segmentation architecture due to its flexibility, optimized modular design, and success in all medical image modalities. Over the years, the U-Net model achieved tremendous attention from academic and industrial researchers. Several extensions of this network have been proposed to address the scale and complexity created by medical tasks. Addressing the deficiency of the naive U-Net model is the foremost step for vendors to utilize the proper U-Net variant model for their business. Having a compendium of different variants in one place makes it easier for builders to identify the relevant research. Also, for ML researchers it will help them understand the challenges of the biological tasks that challenge the model. To address this, we discuss the practical aspects of the U-Net model and suggest a taxonomy to categorize each network variant. Moreover, to measure the performance of these strategies in a clinical application, we propose fair evaluations of some unique and famous designs on well-known datasets. We provide a comprehensive implementation library with trained models for future research. In addition, for ease of future studies, we created an online list of U-Net papers with their possible official implementation. All information is gathered in https://github.com/NITR098/Awesome-U-Net repository.
TransDeepLab: Convolution-Free Transformer-based DeepLab v3+ for Medical Image Segmentation
Aug 01, 2022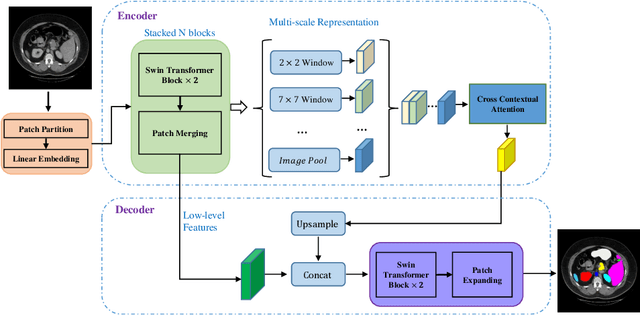
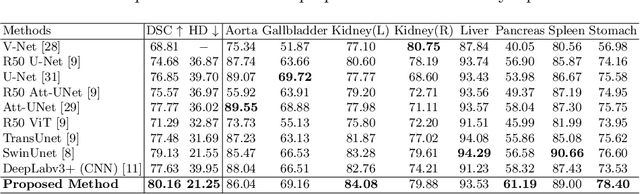
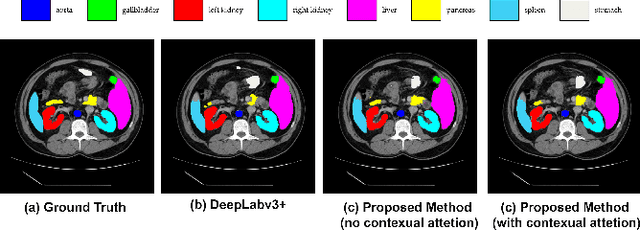
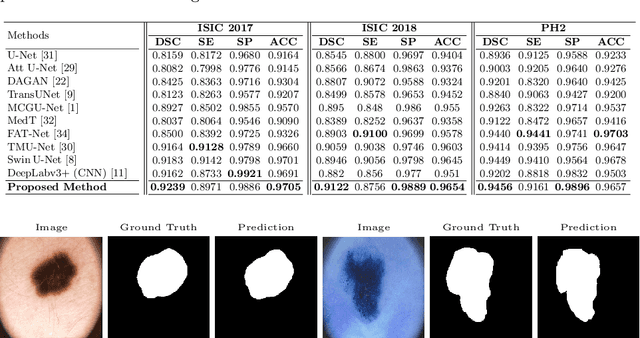
Abstract:Convolutional neural networks (CNNs) have been the de facto standard in a diverse set of computer vision tasks for many years. Especially, deep neural networks based on seminal architectures such as U-shaped models with skip-connections or atrous convolution with pyramid pooling have been tailored to a wide range of medical image analysis tasks. The main advantage of such architectures is that they are prone to detaining versatile local features. However, as a general consensus, CNNs fail to capture long-range dependencies and spatial correlations due to the intrinsic property of confined receptive field size of convolution operations. Alternatively, Transformer, profiting from global information modelling that stems from the self-attention mechanism, has recently attained remarkable performance in natural language processing and computer vision. Nevertheless, previous studies prove that both local and global features are critical for a deep model in dense prediction, such as segmenting complicated structures with disparate shapes and configurations. To this end, this paper proposes TransDeepLab, a novel DeepLab-like pure Transformer for medical image segmentation. Specifically, we exploit hierarchical Swin-Transformer with shifted windows to extend the DeepLabv3 and model the Atrous Spatial Pyramid Pooling (ASPP) module. A thorough search of the relevant literature yielded that we are the first to model the seminal DeepLab model with a pure Transformer-based model. Extensive experiments on various medical image segmentation tasks verify that our approach performs superior or on par with most contemporary works on an amalgamation of Vision Transformer and CNN-based methods, along with a significant reduction of model complexity. The codes and trained models are publicly available at https://github.com/rezazad68/transdeeplab
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge