cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Large Language Model Evaluated Stand-alone Attention-Assisted Graph Neural Network with Spatial and Structural Information Interaction for Precise Endoscopic Image Segmentation
Aug 09, 2025Accurate endoscopic image segmentation on the polyps is critical for early colorectal cancer detection. However, this task remains challenging due to low contrast with surrounding mucosa, specular highlights, and indistinct boundaries. To address these challenges, we propose FOCUS-Med, which stands for Fusion of spatial and structural graph with attentional context-aware polyp segmentation in endoscopic medical imaging. FOCUS-Med integrates a Dual Graph Convolutional Network (Dual-GCN) module to capture contextual spatial and topological structural dependencies. This graph-based representation enables the model to better distinguish polyps from background tissues by leveraging topological cues and spatial connectivity, which are often obscured in raw image intensities. It enhances the model's ability to preserve boundaries and delineate complex shapes typical of polyps. In addition, a location-fused stand-alone self-attention is employed to strengthen global context integration. To bridge the semantic gap between encoder-decoder layers, we incorporate a trainable weighted fast normalized fusion strategy for efficient multi-scale aggregation. Notably, we are the first to introduce the use of a Large Language Model (LLM) to provide detailed qualitative evaluations of segmentation quality. Extensive experiments on public benchmarks demonstrate that FOCUS-Med achieves state-of-the-art performance across five key metrics, underscoring its effectiveness and clinical potential for AI-assisted colonoscopy.
Enhanced Liver Tumor Detection in CT Images Using 3D U-Net and Bat Algorithm for Hyperparameter Optimization
Aug 11, 2025Liver cancer is one of the most prevalent and lethal forms of cancer, making early detection crucial for effective treatment. This paper introduces a novel approach for automated liver tumor segmentation in computed tomography (CT) images by integrating a 3D U-Net architecture with the Bat Algorithm for hyperparameter optimization. The method enhances segmentation accuracy and robustness by intelligently optimizing key parameters like the learning rate and batch size. Evaluated on a publicly available dataset, our model demonstrates a strong ability to balance precision and recall, with a high F1-score at lower prediction thresholds. This is particularly valuable for clinical diagnostics, where ensuring no potential tumors are missed is paramount. Our work contributes to the field of medical image analysis by demonstrating that the synergy between a robust deep learning architecture and a metaheuristic optimization algorithm can yield a highly effective solution for complex segmentation tasks.
Synthetic Data-Driven Multi-Architecture Framework for Automated Polyp Segmentation Through Integrated Detection and Mask Generation
Aug 08, 2025Colonoscopy is a vital tool for the early diagnosis of colorectal cancer, which is one of the main causes of cancer-related mortality globally; hence, it is deemed an essential technique for the prevention and early detection of colorectal cancer. The research introduces a unique multidirectional architectural framework to automate polyp detection within colonoscopy images while helping resolve limited healthcare dataset sizes and annotation complexities. The research implements a comprehensive system that delivers synthetic data generation through Stable Diffusion enhancements together with detection and segmentation algorithms. This detection approach combines Faster R-CNN for initial object localization while the Segment Anything Model (SAM) refines the segmentation masks. The faster R-CNN detection algorithm achieved a recall of 93.08% combined with a precision of 88.97% and an F1 score of 90.98%.SAM is then used to generate the image mask. The research evaluated five state-of-the-art segmentation models that included U-Net, PSPNet, FPN, LinkNet, and MANet using ResNet34 as a base model. The results demonstrate the superior performance of FPN with the highest scores of PSNR (7.205893) and SSIM (0.492381), while UNet excels in recall (84.85%) and LinkNet shows balanced performance in IoU (64.20%) and Dice score (77.53%).
Improving Diagnostic Accuracy for Oral Cancer with inpainting Synthesis Lesions Generated Using Diffusion Models
Aug 08, 2025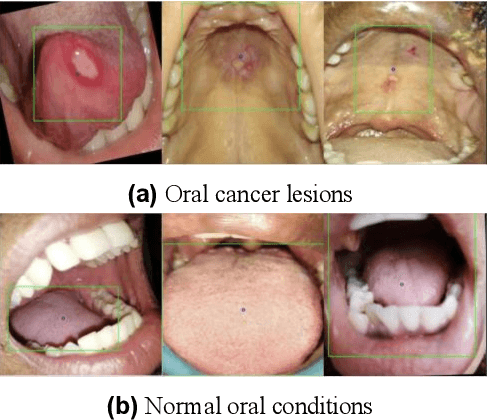
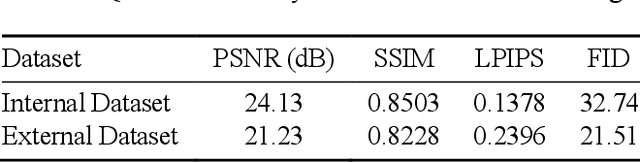
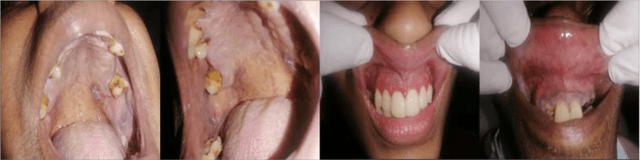
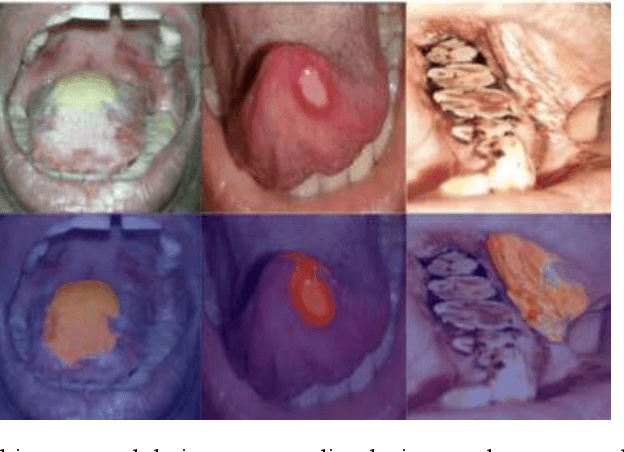
In oral cancer diagnostics, the limited availability of annotated datasets frequently constrains the performance of diagnostic models, particularly due to the variability and insufficiency of training data. To address these challenges, this study proposed a novel approach to enhance diagnostic accuracy by synthesizing realistic oral cancer lesions using an inpainting technique with a fine-tuned diffusion model. We compiled a comprehensive dataset from multiple sources, featuring a variety of oral cancer images. Our method generated synthetic lesions that exhibit a high degree of visual fidelity to actual lesions, thereby significantly enhancing the performance of diagnostic algorithms. The results show that our classification model achieved a diagnostic accuracy of 0.97 in differentiating between cancerous and non-cancerous tissues, while our detection model accurately identified lesion locations with 0.85 accuracy. This method validates the potential for synthetic image generation in medical diagnostics and paves the way for further research into extending these methods to other types of cancer diagnostics.
Model Compression Engine for Wearable Devices Skin Cancer Diagnosis
Jul 23, 2025Skin cancer is one of the most prevalent and preventable types of cancer, yet its early detection remains a challenge, particularly in resource-limited settings where access to specialized healthcare is scarce. This study proposes an AI-driven diagnostic tool optimized for embedded systems to address this gap. Using transfer learning with the MobileNetV2 architecture, the model was adapted for binary classification of skin lesions into "Skin Cancer" and "Other." The TensorRT framework was employed to compress and optimize the model for deployment on the NVIDIA Jetson Orin Nano, balancing performance with energy efficiency. Comprehensive evaluations were conducted across multiple benchmarks, including model size, inference speed, throughput, and power consumption. The optimized models maintained their performance, achieving an F1-Score of 87.18% with a precision of 93.18% and recall of 81.91%. Post-compression results showed reductions in model size of up to 0.41, along with improvements in inference speed and throughput, and a decrease in energy consumption of up to 0.93 in INT8 precision. These findings validate the feasibility of deploying high-performing, energy-efficient diagnostic tools on resource-constrained edge devices. Beyond skin cancer detection, the methodologies applied in this research have broader applications in other medical diagnostics and domains requiring accessible, efficient AI solutions. This study underscores the potential of optimized AI systems to revolutionize healthcare diagnostics, thereby bridging the divide between advanced technology and underserved regions.
Mitigating Multi-Sequence 3D Prostate MRI Data Scarcity through Domain Adaptation using Locally-Trained Latent Diffusion Models for Prostate Cancer Detection
Jul 08, 2025Objective: Latent diffusion models (LDMs) could mitigate data scarcity challenges affecting machine learning development for medical image interpretation. The recent CCELLA LDM improved prostate cancer detection performance using synthetic MRI for classifier training but was limited to the axial T2-weighted (AxT2) sequence, did not investigate inter-institutional domain shift, and prioritized radiology over histopathology outcomes. We propose CCELLA++ to address these limitations and improve clinical utility. Methods: CCELLA++ expands CCELLA for simultaneous biparametric prostate MRI (bpMRI) generation, including the AxT2, high b-value diffusion series (HighB) and apparent diffusion coefficient map (ADC). Domain adaptation was investigated by pretraining classifiers on real or LDM-generated synthetic data from an internal institution, followed with fine-tuning on progressively smaller fractions of an out-of-distribution, external dataset. Results: CCELLA++ improved 3D FID for HighB and ADC but not AxT2 (0.013, 0.012, 0.063 respectively) sequences compared to CCELLA (0.060). Classifier pretraining with CCELLA++ bpMRI outperformed real bpMRI in AP and AUC for all domain adaptation scenarios. CCELLA++ pretraining achieved highest classifier performance below 50% (n=665) external dataset volume. Conclusion: Synthetic bpMRI generated by our method can improve downstream classifier generalization and performance beyond real bpMRI or CCELLA-generated AxT2-only images. Future work should seek to quantify medical image sample quality, balance multi-sequence LDM training, and condition the LDM with additional information. Significance: The proposed CCELLA++ LDM can generate synthetic bpMRI that outperforms real data for domain adaptation with a limited target institution dataset. Our code is available at https://github.com/grabkeem/CCELLA-plus-plus
A Hybrid CNN-VSSM model for Multi-View, Multi-Task Mammography Analysis: Robust Diagnosis with Attention-Based Fusion
Jul 22, 2025Early and accurate interpretation of screening mammograms is essential for effective breast cancer detection, yet it remains a complex challenge due to subtle imaging findings and diagnostic ambiguity. Many existing AI approaches fall short by focusing on single view inputs or single-task outputs, limiting their clinical utility. To address these limitations, we propose a novel multi-view, multitask hybrid deep learning framework that processes all four standard mammography views and jointly predicts diagnostic labels and BI-RADS scores for each breast. Our architecture integrates a hybrid CNN VSSM backbone, combining convolutional encoders for rich local feature extraction with Visual State Space Models (VSSMs) to capture global contextual dependencies. To improve robustness and interpretability, we incorporate a gated attention-based fusion module that dynamically weights information across views, effectively handling cases with missing data. We conduct extensive experiments across diagnostic tasks of varying complexity, benchmarking our proposed hybrid models against baseline CNN architectures and VSSM models in both single task and multi task learning settings. Across all tasks, the hybrid models consistently outperform the baselines. In the binary BI-RADS 1 vs. 5 classification task, the shared hybrid model achieves an AUC of 0.9967 and an F1 score of 0.9830. For the more challenging ternary classification, it attains an F1 score of 0.7790, while in the five-class BI-RADS task, the best F1 score reaches 0.4904. These results highlight the effectiveness of the proposed hybrid framework and underscore both the potential and limitations of multitask learning for improving diagnostic performance and enabling clinically meaningful mammography analysis.
MyGO: Make your Goals Obvious, Avoiding Semantic Confusion in Prostate Cancer Lesion Region Segmentation
Jul 23, 2025Early diagnosis and accurate identification of lesion location and progression in prostate cancer (PCa) are critical for assisting clinicians in formulating effective treatment strategies. However, due to the high semantic homogeneity between lesion and non-lesion areas, existing medical image segmentation methods often struggle to accurately comprehend lesion semantics, resulting in the problem of semantic confusion. To address this challenge, we propose a novel Pixel Anchor Module, which guides the model to discover a sparse set of feature anchors that serve to capture and interpret global contextual information. This mechanism enhances the model's nonlinear representation capacity and improves segmentation accuracy within lesion regions. Moreover, we design a self-attention-based Top_k selection strategy to further refine the identification of these feature anchors, and incorporate a focal loss function to mitigate class imbalance, thereby facilitating more precise semantic interpretation across diverse regions. Our method achieves state-of-the-art performance on the PI-CAI dataset, demonstrating 69.73% IoU and 74.32% Dice scores, and significantly improving prostate cancer lesion detection.
Towards Human-AI Collaboration System for the Detection of Invasive Ductal Carcinoma in Histopathology Images
Aug 11, 2025Invasive ductal carcinoma (IDC) is the most prevalent form of breast cancer, and early, accurate diagnosis is critical to improving patient survival rates by guiding treatment decisions. Combining medical expertise with artificial intelligence (AI) holds significant promise for enhancing the precision and efficiency of IDC detection. In this work, we propose a human-in-the-loop (HITL) deep learning system designed to detect IDC in histopathology images. The system begins with an initial diagnosis provided by a high-performance EfficientNetV2S model, offering feedback from AI to the human expert. Medical professionals then review the AI-generated results, correct any misclassified images, and integrate the revised labels into the training dataset, forming a feedback loop from the human back to the AI. This iterative process refines the model's performance over time. The EfficientNetV2S model itself achieves state-of-the-art performance compared to existing methods in the literature, with an overall accuracy of 93.65\%. Incorporating the human-in-the-loop system further improves the model's accuracy using four experimental groups with misclassified images. These results demonstrate the potential of this collaborative approach to enhance AI performance in diagnostic systems. This work contributes to advancing automated, efficient, and highly accurate methods for IDC detection through human-AI collaboration, offering a promising direction for future AI-assisted medical diagnostics.
A computationally frugal open-source foundation model for thoracic disease detection in lung cancer screening programs
Jul 02, 2025Low-dose computed tomography (LDCT) imaging employed in lung cancer screening (LCS) programs is increasing in uptake worldwide. LCS programs herald a generational opportunity to simultaneously detect cancer and non-cancer-related early-stage lung disease. Yet these efforts are hampered by a shortage of radiologists to interpret scans at scale. Here, we present TANGERINE, a computationally frugal, open-source vision foundation model for volumetric LDCT analysis. Designed for broad accessibility and rapid adaptation, TANGERINE can be fine-tuned off the shelf for a wide range of disease-specific tasks with limited computational resources and training data. Relative to models trained from scratch, TANGERINE demonstrates fast convergence during fine-tuning, thereby requiring significantly fewer GPU hours, and displays strong label efficiency, achieving comparable or superior performance with a fraction of fine-tuning data. Pretrained using self-supervised learning on over 98,000 thoracic LDCTs, including the UK's largest LCS initiative to date and 27 public datasets, TANGERINE achieves state-of-the-art performance across 14 disease classification tasks, including lung cancer and multiple respiratory diseases, while generalising robustly across diverse clinical centres. By extending a masked autoencoder framework to 3D imaging, TANGERINE offers a scalable solution for LDCT analysis, departing from recent closed, resource-intensive models by combining architectural simplicity, public availability, and modest computational requirements. Its accessible, open-source lightweight design lays the foundation for rapid integration into next-generation medical imaging tools that could transform LCS initiatives, allowing them to pivot from a singular focus on lung cancer detection to comprehensive respiratory disease management in high-risk populations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge