cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Graph Kolmogorov-Arnold Networks for Multi-Cancer Classification and Biomarker Identification, An Interpretable Multi-Omics Approach
Mar 29, 2025The integration of multi-omics data presents a major challenge in precision medicine, requiring advanced computational methods for accurate disease classification and biological interpretation. This study introduces the Multi-Omics Graph Kolmogorov-Arnold Network (MOGKAN), a deep learning model that integrates messenger RNA, micro RNA sequences, and DNA methylation data with Protein-Protein Interaction (PPI) networks for accurate and interpretable cancer classification across 31 cancer types. MOGKAN employs a hybrid approach combining differential expression with DESeq2, Linear Models for Microarray (LIMMA), and Least Absolute Shrinkage and Selection Operator (LASSO) regression to reduce multi-omics data dimensionality while preserving relevant biological features. The model architecture is based on the Kolmogorov-Arnold theorem principle, using trainable univariate functions to enhance interpretability and feature analysis. MOGKAN achieves classification accuracy of 96.28 percent and demonstrates low experimental variability with a standard deviation that is reduced by 1.58 to 7.30 percents compared to Convolutional Neural Networks (CNNs) and Graph Neural Networks (GNNs). The biomarkers identified by MOGKAN have been validated as cancer-related markers through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. The proposed model presents an ability to uncover molecular oncogenesis mechanisms by detecting phosphoinositide-binding substances and regulating sphingolipid cellular processes. By integrating multi-omics data with graph-based deep learning, our proposed approach demonstrates superior predictive performance and interpretability that has the potential to enhance the translation of complex multi-omics data into clinically actionable cancer diagnostics.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025



Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
Single Shot AI-assisted quantification of KI-67 proliferation index in breast cancer
Mar 25, 2025Reliable quantification of Ki-67, a key proliferation marker in breast cancer, is essential for molecular subtyping and informed treatment planning. Conventional approaches, including visual estimation and manual counting, suffer from interobserver variability and limited reproducibility. This study introduces an AI-assisted method using the YOLOv8 object detection framework for automated Ki-67 scoring. High-resolution digital images (40x magnification) of immunohistochemically stained tumor sections were captured from Ki-67 hotspot regions and manually annotated by a domain expert to distinguish Ki-67-positive and negative tumor cells. The dataset was augmented and divided into training (80%), validation (10%), and testing (10%) subsets. Among the YOLOv8 variants tested, the Medium model achieved the highest performance, with a mean Average Precision at 50% Intersection over Union (mAP50) exceeding 85% for Ki-67-positive cells. The proposed approach offers an efficient, scalable, and objective alternative to conventional scoring methods, supporting greater consistency in Ki-67 evaluation. Future directions include developing user-friendly clinical interfaces and expanding to multi-institutional datasets to enhance generalizability and facilitate broader adoption in diagnostic practice.
Tumor monitoring and detection of lymph node metastasis using quantitative ultrasound and immune cytokine profiling in dogs undergoing radiation therapy: a pilot study
Mar 25, 2025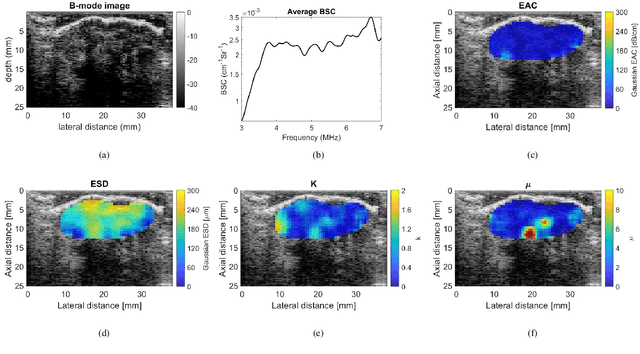
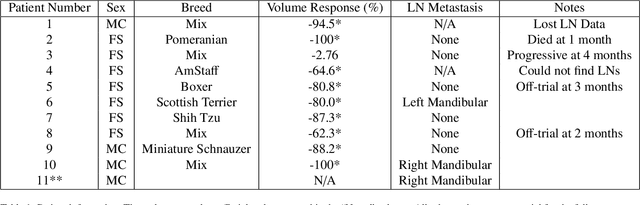
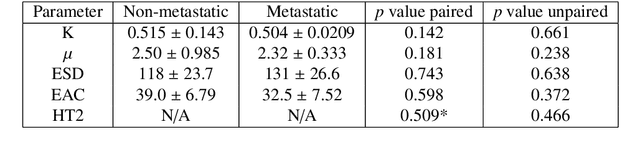
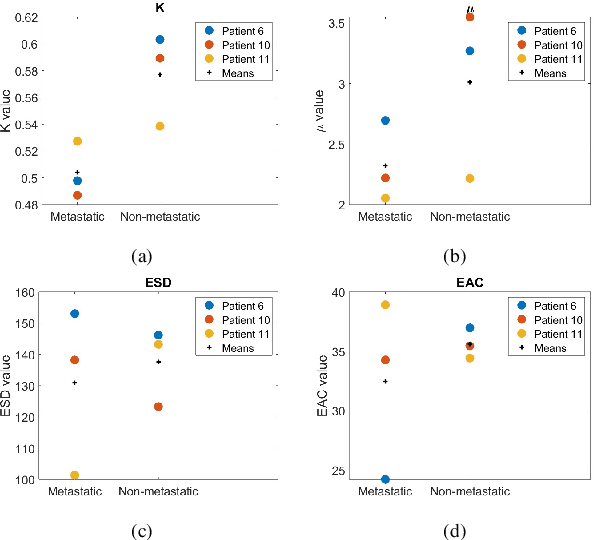
Quantitative ultrasound (QUS) characterizes the composition of cells to distinguish diseased from healthy tissue. QUS can reflect the complexity of the tumor and detect early lymph node (LN) metastasis ex vivo. The objective in this study was to gather preliminary QUS and cytokine data from dogs undergoing radiation therapy and correlate QUS data with both LN metastasis and tumor response. Spontaneous solid tumors were evaluated with QUS before and up to one year after receiving RT. Additionally, regional LNs were evaluated with QUS in vivo, then excised and examined with histopathology to detect metastasis. Paired t-tests were used to compare QUS data of metastatic and non-metastatic LNs within patients. Furthermore, paired t-tests compared pre- versus post-RT QUS data. Serum was collected at each time point for cytokine profiles. Most statistical tests were underpowered to produce significant p values, but interesting trends were observed. The lowest p values for LN tests were found with the envelope statistics K (p = 0.142) and mu (p = 0.181), which correspond to cell structure and number of scatterers. For tumor response, the lowest p values were found with K (p = 0.115) and mu (p = 0.127) when comparing baseline QUS data with QUS data 1 week after RT. Monocyte chemoattractant protein 1 (MCP-1) was significantly higher in dogs with cancer when compared to healthy controls (p = 1.12e-4). A weak correlation was found between effective scatterer diameter (ESD) and Transforming growth factor beta 1 (TGFB-1). While statistical tests on the preliminary QUS data alone were underpowered to detect significant differences among groups, our methods create a basis for future studies.
SMILE: a Scale-aware Multiple Instance Learning Method for Multicenter STAS Lung Cancer Histopathology Diagnosis
Mar 18, 2025Spread through air spaces (STAS) represents a newly identified aggressive pattern in lung cancer, which is known to be associated with adverse prognostic factors and complex pathological features. Pathologists currently rely on time consuming manual assessments, which are highly subjective and prone to variation. This highlights the urgent need for automated and precise diag nostic solutions. 2,970 lung cancer tissue slides are comprised from multiple centers, re-diagnosed them, and constructed and publicly released three lung cancer STAS datasets: STAS CSU (hospital), STAS TCGA, and STAS CPTAC. All STAS datasets provide corresponding pathological feature diagnoses and related clinical data. To address the bias, sparse and heterogeneous nature of STAS, we propose an scale-aware multiple instance learning(SMILE) method for STAS diagnosis of lung cancer. By introducing a scale-adaptive attention mechanism, the SMILE can adaptively adjust high attention instances, reducing over-reliance on local regions and promoting consistent detection of STAS lesions. Extensive experiments show that SMILE achieved competitive diagnostic results on STAS CSU, diagnosing 251 and 319 STAS samples in CPTAC andTCGA,respectively, surpassing clinical average AUC. The 11 open baseline results are the first to be established for STAS research, laying the foundation for the future expansion, interpretability, and clinical integration of computational pathology technologies. The datasets and code are available at https://anonymous.4open.science/r/IJCAI25-1DA1.
Leveraging Sparse Annotations for Leukemia Diagnosis on the Large Leukemia Dataset
Apr 03, 2025Leukemia is 10th most frequently diagnosed cancer and one of the leading causes of cancer related deaths worldwide. Realistic analysis of Leukemia requires White Blook Cells (WBC) localization, classification, and morphological assessment. Despite deep learning advances in medical imaging, leukemia analysis lacks a large, diverse multi-task dataset, while existing small datasets lack domain diversity, limiting real world applicability. To overcome dataset challenges, we present a large scale WBC dataset named Large Leukemia Dataset (LLD) and novel methods for detecting WBC with their attributes. Our contribution here is threefold. First, we present a large-scale Leukemia dataset collected through Peripheral Blood Films (PBF) from several patients, through multiple microscopes, multi cameras, and multi magnification. To enhance diagnosis explainability and medical expert acceptance, each leukemia cell is annotated at 100x with 7 morphological attributes, ranging from Cell Size to Nuclear Shape. Secondly, we propose a multi task model that not only detects WBCs but also predicts their attributes, providing an interpretable and clinically meaningful solution. Third, we propose a method for WBC detection with attribute analysis using sparse annotations. This approach reduces the annotation burden on hematologists, requiring them to mark only a small area within the field of view. Our method enables the model to leverage the entire field of view rather than just the annotated regions, enhancing learning efficiency and diagnostic accuracy. From diagnosis explainability to overcoming domain shift challenges, presented datasets could be used for many challenging aspects of microscopic image analysis. The datasets, code, and demo are available at: https://im.itu.edu.pk/sparse-leukemiaattri/
Efficient Brain Tumor Segmentation Using a Dual-Decoder 3D U-Net with Attention Gates (DDUNet)
Apr 14, 2025



Cancer remains one of the leading causes of mortality worldwide, and among its many forms, brain tumors are particularly notorious due to their aggressive nature and the critical challenges involved in early diagnosis. Recent advances in artificial intelligence have shown great promise in assisting medical professionals with precise tumor segmentation, a key step in timely diagnosis and treatment planning. However, many state-of-the-art segmentation methods require extensive computational resources and prolonged training times, limiting their practical application in resource-constrained settings. In this work, we present a novel dual-decoder U-Net architecture enhanced with attention-gated skip connections, designed specifically for brain tumor segmentation from MRI scans. Our approach balances efficiency and accuracy by achieving competitive segmentation performance while significantly reducing training demands. Evaluated on the BraTS 2020 dataset, the proposed model achieved Dice scores of 85.06% for Whole Tumor (WT), 80.61% for Tumor Core (TC), and 71.26% for Enhancing Tumor (ET) in only 50 epochs, surpassing several commonly used U-Net variants. Our model demonstrates that high-quality brain tumor segmentation is attainable even under limited computational resources, thereby offering a viable solution for researchers and clinicians operating with modest hardware. This resource-efficient model has the potential to improve early detection and diagnosis of brain tumors, ultimately contributing to better patient outcomes
How Good is my Histopathology Vision-Language Foundation Model? A Holistic Benchmark
Mar 17, 2025



Recently, histopathology vision-language foundation models (VLMs) have gained popularity due to their enhanced performance and generalizability across different downstream tasks. However, most existing histopathology benchmarks are either unimodal or limited in terms of diversity of clinical tasks, organs, and acquisition instruments, as well as their partial availability to the public due to patient data privacy. As a consequence, there is a lack of comprehensive evaluation of existing histopathology VLMs on a unified benchmark setting that better reflects a wide range of clinical scenarios. To address this gap, we introduce HistoVL, a fully open-source comprehensive benchmark comprising images acquired using up to 11 various acquisition tools that are paired with specifically crafted captions by incorporating class names and diverse pathology descriptions. Our Histo-VL includes 26 organs, 31 cancer types, and a wide variety of tissue obtained from 14 heterogeneous patient cohorts, totaling more than 5 million patches obtained from over 41K WSIs viewed under various magnification levels. We systematically evaluate existing histopathology VLMs on Histo-VL to simulate diverse tasks performed by experts in real-world clinical scenarios. Our analysis reveals interesting findings, including large sensitivity of most existing histopathology VLMs to textual changes with a drop in balanced accuracy of up to 25% in tasks such as Metastasis detection, low robustness to adversarial attacks, as well as improper calibration of models evident through high ECE values and low model prediction confidence, all of which can affect their clinical implementation.
SCFANet: Style Distribution Constraint Feature Alignment Network For Pathological Staining Translation
Apr 01, 2025



Immunohistochemical (IHC) staining serves as a valuable technique for detecting specific antigens or proteins through antibody-mediated visualization. However, the IHC staining process is both time-consuming and costly. To address these limitations, the application of deep learning models for direct translation of cost-effective Hematoxylin and Eosin (H&E) stained images into IHC stained images has emerged as an efficient solution. Nevertheless, the conversion from H&E to IHC images presents significant challenges, primarily due to alignment discrepancies between image pairs and the inherent diversity in IHC staining style patterns. To overcome these challenges, we propose the Style Distribution Constraint Feature Alignment Network (SCFANet), which incorporates two innovative modules: the Style Distribution Constrainer (SDC) and Feature Alignment Learning (FAL). The SDC ensures consistency between the generated and target images' style distributions while integrating cycle consistency loss to maintain structural consistency. To mitigate the complexity of direct image-to-image translation, the FAL module decomposes the end-to-end translation task into two subtasks: image reconstruction and feature alignment. Furthermore, we ensure pathological consistency between generated and target images by maintaining pathological pattern consistency and Optical Density (OD) uniformity. Extensive experiments conducted on the Breast Cancer Immunohistochemical (BCI) dataset demonstrate that our SCFANet model outperforms existing methods, achieving precise transformation of H&E-stained images into their IHC-stained counterparts. The proposed approach not only addresses the technical challenges in H&E to IHC image translation but also provides a robust framework for accurate and efficient stain conversion in pathological analysis.
SYN-LUNGS: Towards Simulating Lung Nodules with Anatomy-Informed Digital Twins for AI Training
Feb 28, 2025



AI models for lung cancer screening are limited by data scarcity, impacting generalizability and clinical applicability. Generative models address this issue but are constrained by training data variability. We introduce SYN-LUNGS, a framework for generating high-quality 3D CT images with detailed annotations. SYN-LUNGS integrates XCAT3 phantoms for digital twin generation, X-Lesions for nodule simulation (varying size, location, and appearance), and DukeSim for CT image formation with vendor and parameter variability. The dataset includes 3,072 nodule images from 1,044 simulated CT scans, with 512 lesions and 174 digital twins. Models trained on clinical + simulated data outperform clinical only models, achieving 10% improvement in detection, 2-9% in segmentation and classification, and enhanced synthesis.By incorporating anatomy-informed simulations, SYN-LUNGS provides a scalable approach for AI model development, particularly in rare disease representation and improving model reliability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge