cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Transfer Learning and Explainable AI for Brain Tumor Classification: A Study Using MRI Data from Bangladesh
Jun 08, 2025Brain tumors, regardless of being benign or malignant, pose considerable health risks, with malignant tumors being more perilous due to their swift and uncontrolled proliferation, resulting in malignancy. Timely identification is crucial for enhancing patient outcomes, particularly in nations such as Bangladesh, where healthcare infrastructure is constrained. Manual MRI analysis is arduous and susceptible to inaccuracies, rendering it inefficient for prompt diagnosis. This research sought to tackle these problems by creating an automated brain tumor classification system utilizing MRI data obtained from many hospitals in Bangladesh. Advanced deep learning models, including VGG16, VGG19, and ResNet50, were utilized to classify glioma, meningioma, and various brain cancers. Explainable AI (XAI) methodologies, such as Grad-CAM and Grad-CAM++, were employed to improve model interpretability by emphasizing the critical areas in MRI scans that influenced the categorization. VGG16 achieved the most accuracy, attaining 99.17%. The integration of XAI enhanced the system's transparency and stability, rendering it more appropriate for clinical application in resource-limited environments such as Bangladesh. This study highlights the capability of deep learning models, in conjunction with explainable artificial intelligence (XAI), to enhance brain tumor detection and identification in areas with restricted access to advanced medical technologies.
MERA: Multimodal and Multiscale Self-Explanatory Model with Considerably Reduced Annotation for Lung Nodule Diagnosis
Apr 27, 2025



Lung cancer, a leading cause of cancer-related deaths globally, emphasises the importance of early detection for better patient outcomes. Pulmonary nodules, often early indicators of lung cancer, necessitate accurate, timely diagnosis. Despite Explainable Artificial Intelligence (XAI) advances, many existing systems struggle providing clear, comprehensive explanations, especially with limited labelled data. This study introduces MERA, a Multimodal and Multiscale self-Explanatory model designed for lung nodule diagnosis with considerably Reduced Annotation requirements. MERA integrates unsupervised and weakly supervised learning strategies (self-supervised learning techniques and Vision Transformer architecture for unsupervised feature extraction) and a hierarchical prediction mechanism leveraging sparse annotations via semi-supervised active learning in the learned latent space. MERA explains its decisions on multiple levels: model-level global explanations via semantic latent space clustering, instance-level case-based explanations showing similar instances, local visual explanations via attention maps, and concept explanations using critical nodule attributes. Evaluations on the public LIDC dataset show MERA's superior diagnostic accuracy and self-explainability. With only 1% annotated samples, MERA achieves diagnostic accuracy comparable to or exceeding state-of-the-art methods requiring full annotation. The model's inherent design delivers comprehensive, robust, multilevel explanations aligned closely with clinical practice, enhancing trustworthiness and transparency. Demonstrated viability of unsupervised and weakly supervised learning lowers the barrier to deploying diagnostic AI in broader medical domains. Our complete code is open-source available: https://github.com/diku-dk/credanno.
Adaptive Deep Learning for Multiclass Breast Cancer Classification via Misprediction Risk Analysis
Mar 17, 2025



Breast cancer remains one of the leading causes of cancer-related deaths worldwide. Early detection is crucial for improving patient outcomes, yet the diagnostic process is often complex and prone to inconsistencies among pathologists. Computer-aided diagnostic approaches have significantly enhanced breast cancer detection, particularly in binary classification (benign vs. malignant). However, these methods face challenges in multiclass classification, leading to frequent mispredictions. In this work, we propose a novel adaptive learning approach for multiclass breast cancer classification using H&E-stained histopathology images. First, we introduce a misprediction risk analysis framework that quantifies and ranks the likelihood of an image being mislabeled by a classifier. This framework leverages an interpretable risk model that requires only a small number of labeled samples for training. Next, we present an adaptive learning strategy that fine-tunes classifiers based on the specific characteristics of a given dataset. This approach minimizes misprediction risk, allowing the classifier to adapt effectively to the target workload. We evaluate our proposed solutions on real benchmark datasets, demonstrating that our risk analysis framework more accurately identifies mispredictions compared to existing methods. Furthermore, our adaptive learning approach significantly improves the performance of state-of-the-art deep neural network classifiers.
CPLOYO: A Pulmonary Nodule Detection Model with Multi-Scale Feature Fusion and Nonlinear Feature Learning
Mar 13, 2025



The integration of Internet of Things (IoT) technology in pulmonary nodule detection significantly enhances the intelligence and real-time capabilities of the detection system. Currently, lung nodule detection primarily focuses on the identification of solid nodules, but different types of lung nodules correspond to various forms of lung cancer. Multi-type detection contributes to improving the overall lung cancer detection rate and enhancing the cure rate. To achieve high sensitivity in nodule detection, targeted improvements were made to the YOLOv8 model. Firstly, the C2f\_RepViTCAMF module was introduced to augment the C2f module in the backbone, thereby enhancing detection accuracy for small lung nodules and achieving a lightweight model design. Secondly, the MSCAF module was incorporated to reconstruct the feature fusion section of the model, improving detection accuracy for lung nodules of varying scales. Furthermore, the KAN network was integrated into the model. By leveraging the KAN network's powerful nonlinear feature learning capability, detection accuracy for small lung nodules was further improved, and the model's generalization ability was enhanced. Tests conducted on the LUNA16 dataset demonstrate that the improved model outperforms the original model as well as other mainstream models such as YOLOv9 and RT-DETR across various evaluation metrics.
Multimodal AI-driven Biomarker for Early Detection of Cancer Cachexia
Mar 09, 2025Cancer cachexia is a multifactorial syndrome characterized by progressive muscle wasting, metabolic dysfunction, and systemic inflammation, leading to reduced quality of life and increased mortality. Despite extensive research, no single definitive biomarker exists, as cachexia-related indicators such as serum biomarkers, skeletal muscle measurements, and metabolic abnormalities often overlap with other conditions. Existing composite indices, including the Cancer Cachexia Index (CXI), Modified CXI (mCXI), and Cachexia Score (CASCO), integrate multiple biomarkers but lack standardized thresholds, limiting their clinical utility. This study proposes a multimodal AI-based biomarker for early cancer cachexia detection, leveraging open-source large language models (LLMs) and foundation models trained on medical data. The approach integrates heterogeneous patient data, including demographics, disease status, lab reports, radiological imaging (CT scans), and clinical notes, using a machine learning framework that can handle missing data. Unlike previous AI-based models trained on curated datasets, this method utilizes routinely collected clinical data, enhancing real-world applicability. Additionally, the model incorporates confidence estimation, allowing the identification of cases requiring expert review for precise clinical interpretation. Preliminary findings demonstrate that integrating multiple data modalities improves cachexia prediction accuracy at the time of cancer diagnosis. The AI-based biomarker dynamically adapts to patient-specific factors such as age, race, ethnicity, weight, cancer type, and stage, avoiding the limitations of fixed-threshold biomarkers. This multimodal AI biomarker provides a scalable and clinically viable solution for early cancer cachexia detection, facilitating personalized interventions and potentially improving treatment outcomes and patient survival.
Advancements in Real-Time Oncology Diagnosis: Harnessing AI and Image Fusion Techniques
Mar 14, 2025Real-time computer-aided diagnosis using artificial intelligence (AI), with images, can help oncologists diagnose cancer with high accuracy and in an early phase. We reviewed real-time AI-based analyzed images for decision-making in different cancer types. This paper provides insights into the present and future potential of real-time imaging and image fusion. It explores various real-time techniques, encompassing technical solutions, AI-based imaging, and image fusion diagnosis across multiple anatomical areas, and electromagnetic needle tracking. To provide a thorough overview, this paper discusses ultrasound image fusion, real-time in vivo cancer diagnosis with different spectroscopic techniques, different real-time optical imaging-based cancer diagnosis techniques, elastography-based cancer diagnosis, cervical cancer detection using neuromorphic architectures, different fluorescence image-based cancer diagnosis techniques, and hyperspectral imaging-based cancer diagnosis. We close by offering a more futuristic overview to solve existing problems in real-time image-based cancer diagnosis.
Improving the generalization of deep learning models in the segmentation of mammography images
Mar 28, 2025



Mammography stands as the main screening method for detecting breast cancer early, enhancing treatment success rates. The segmentation of landmark structures in mammography images can aid the medical assessment in the evaluation of cancer risk and the image acquisition adequacy. We introduce a series of data-centric strategies aimed at enriching the training data for deep learning-based segmentation of landmark structures. Our approach involves augmenting the training samples through annotation-guided image intensity manipulation and style transfer to achieve better generalization than standard training procedures. These augmentations are applied in a balanced manner to ensure the model learns to process a diverse range of images generated by different vendor equipments while retaining its efficacy on the original data. We present extensive numerical and visual results that demonstrate the superior generalization capabilities of our methods when compared to the standard training. For this evaluation, we consider a large dataset that includes mammography images generated by different vendor equipments. Further, we present complementary results that show both the strengths and limitations of our methods across various scenarios. The accuracy and robustness demonstrated in the experiments suggest that our method is well-suited for integration into clinical practice.
ColonScopeX: Leveraging Explainable Expert Systems with Multimodal Data for Improved Early Diagnosis of Colorectal Cancer
Apr 09, 2025Colorectal cancer (CRC) ranks as the second leading cause of cancer-related deaths and the third most prevalent malignant tumour worldwide. Early detection of CRC remains problematic due to its non-specific and often embarrassing symptoms, which patients frequently overlook or hesitate to report to clinicians. Crucially, the stage at which CRC is diagnosed significantly impacts survivability, with a survival rate of 80-95\% for Stage I and a stark decline to 10\% for Stage IV. Unfortunately, in the UK, only 14.4\% of cases are diagnosed at the earliest stage (Stage I). In this study, we propose ColonScopeX, a machine learning framework utilizing explainable AI (XAI) methodologies to enhance the early detection of CRC and pre-cancerous lesions. Our approach employs a multimodal model that integrates signals from blood sample measurements, processed using the Savitzky-Golay algorithm for fingerprint smoothing, alongside comprehensive patient metadata, including medication history, comorbidities, age, weight, and BMI. By leveraging XAI techniques, we aim to render the model's decision-making process transparent and interpretable, thereby fostering greater trust and understanding in its predictions. The proposed framework could be utilised as a triage tool or a screening tool of the general population. This research highlights the potential of combining diverse patient data sources and explainable machine learning to tackle critical challenges in medical diagnostics.
Anomaly-Driven Approach for Enhanced Prostate Cancer Segmentation
Apr 30, 2025



Magnetic Resonance Imaging (MRI) plays an important role in identifying clinically significant prostate cancer (csPCa), yet automated methods face challenges such as data imbalance, variable tumor sizes, and a lack of annotated data. This study introduces Anomaly-Driven U-Net (adU-Net), which incorporates anomaly maps derived from biparametric MRI sequences into a deep learning-based segmentation framework to improve csPCa identification. We conduct a comparative analysis of anomaly detection methods and evaluate the integration of anomaly maps into the segmentation pipeline. Anomaly maps, generated using Fixed-Point GAN reconstruction, highlight deviations from normal prostate tissue, guiding the segmentation model to potential cancerous regions. We compare the performance by using the average score, computed as the mean of the AUROC and Average Precision (AP). On the external test set, adU-Net achieves the best average score of 0.618, outperforming the baseline nnU-Net model (0.605). The results demonstrate that incorporating anomaly detection into segmentation improves generalization and performance, particularly with ADC-based anomaly maps, offering a promising direction for automated csPCa identification.
Distributed U-net model and Image Segmentation for Lung Cancer Detection
Feb 20, 2025
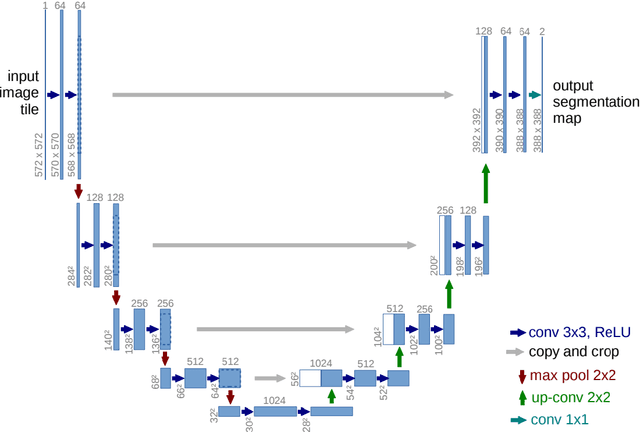
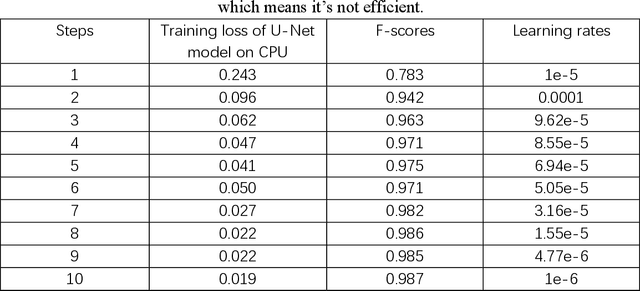
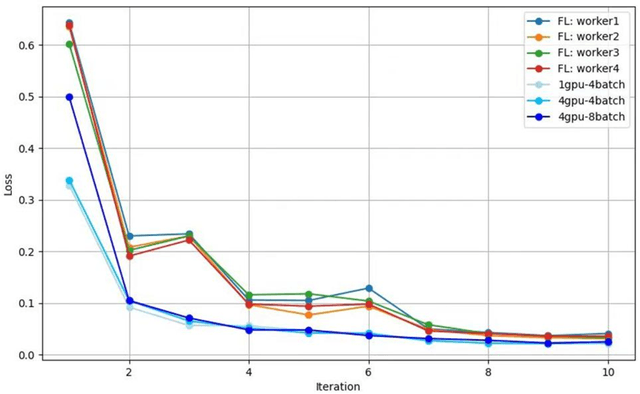
Until now, in the wake of the COVID-19 pandemic in 2019, lung diseases, especially diseases such as lung cancer and chronic obstructive pulmonary disease (COPD), have become an urgent global health issue. In order to mitigate the goal problem, early detection and accurate diagnosis of these conditions are critical for effective treatment and improved patient outcomes. To further research and reduce the error rate of hospital diagnoses, this comprehensive study explored the potential of computer-aided design (CAD) systems, especially utilizing advanced deep learning models such as U-Net. And compared with the literature content of other authors, this study explores the capabilities of U-Net in detail, and enhances the ability to simulate CAD systems through the VGG16 algorithm. An extensive dataset consisting of lung CT images and corresponding segmentation masks, curated collaboratively by multiple academic institutions, serves as the basis for empirical validation. In this paper, the efficiency of U-Net model is evaluated rigorously and precisely under multiple hardware configurations, such as single CPU, single GPU, distributed GPU and federated learning, and the effectiveness and development of the method in the segmentation task of lung disease are demonstrated. Empirical results clearly affirm the robust performance of the U-Net model, most effectively utilizing four GPUs for distributed learning, and these results highlight the potential of U-Net-based CAD systems for accurate and timely lung disease detection and diagnosis huge potential.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge