Zuo Wang
A Novel Graph-Sequence Learning Model for Inductive Text Classification
Dec 23, 2025Abstract:Text classification plays an important role in various downstream text-related tasks, such as sentiment analysis, fake news detection, and public opinion analysis. Recently, text classification based on Graph Neural Networks (GNNs) has made significant progress due to their strong capabilities of structural relationship learning. However, these approaches still face two major limitations. First, these approaches fail to fully consider the diverse structural information across word pairs, e.g., co-occurrence, syntax, and semantics. Furthermore, they neglect sequence information in the text graph structure information learning module and can not classify texts with new words and relations. In this paper, we propose a Novel Graph-Sequence Learning Model for Inductive Text Classification (TextGSL) to address the previously mentioned issues. More specifically, we construct a single text-level graph for all words in each text and establish different edge types based on the diverse relationships between word pairs. Building upon this, we design an adaptive multi-edge message-passing paradigm to aggregate diverse structural information between word pairs. Additionally, sequential information among text data can be captured by the proposed TextGSL through the incorporation of Transformer layers. Therefore, TextGSL can learn more discriminative text representations. TextGSL has been comprehensively compared with several strong baselines. The experimental results on diverse benchmarking datasets demonstrate that TextGSL outperforms these baselines in terms of accuracy.
Jensen-Shannon Divergence Message-Passing for Rich-Text Graph Representation Learning
Dec 23, 2025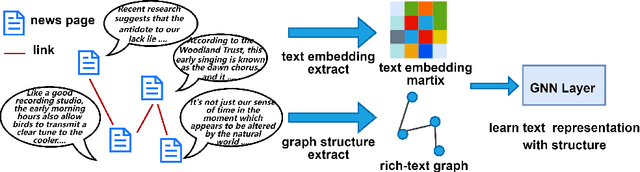
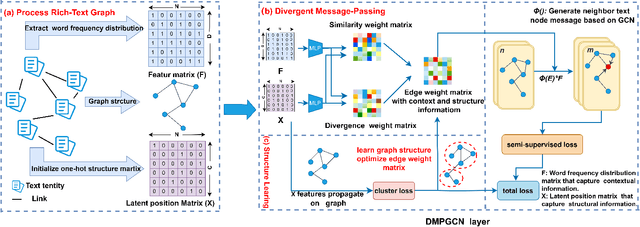
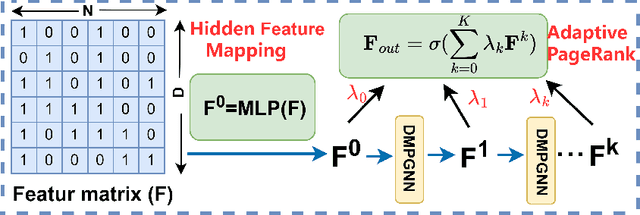
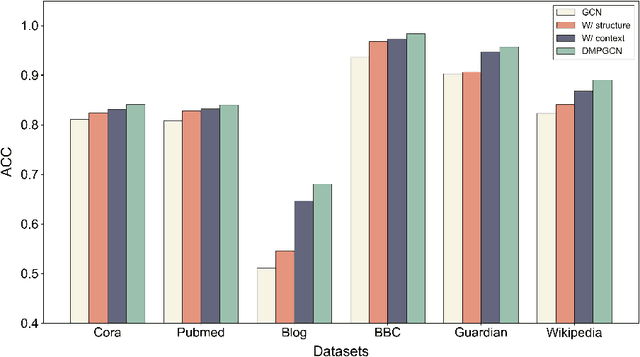
Abstract:In this paper, we investigate how the widely existing contextual and structural divergence may influence the representation learning in rich-text graphs. To this end, we propose Jensen-Shannon Divergence Message-Passing (JSDMP), a new learning paradigm for rich-text graph representation learning. Besides considering similarity regarding structure and text, JSDMP further captures their corresponding dissimilarity by Jensen-Shannon divergence. Similarity and dissimilarity are then jointly used to compute new message weights among text nodes, thus enabling representations to learn with contextual and structural information from truly correlated text nodes. With JSDMP, we propose two novel graph neural networks, namely Divergent message-passing graph convolutional network (DMPGCN) and Divergent message-passing Page-Rank graph neural networks (DMPPRG), for learning representations in rich-text graphs. DMPGCN and DMPPRG have been extensively texted on well-established rich-text datasets and compared with several state-of-the-art baselines. The experimental results show that DMPGCN and DMPPRG can outperform other baselines, demonstrating the effectiveness of the proposed Jensen-Shannon Divergence Message-Passing paradigm
Causal Heterogeneous Graph Learning Method for Chronic Obstructive Pulmonary Disease Prediction
Dec 22, 2025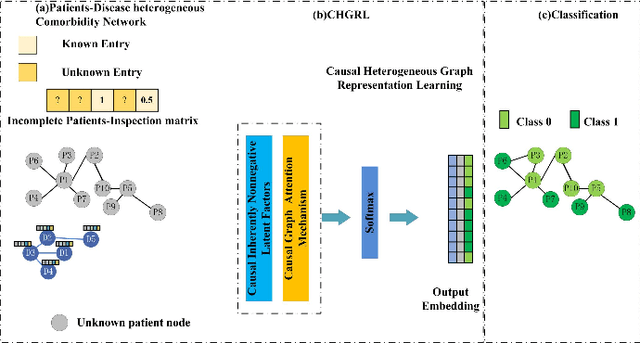
Abstract:Due to the insufficient diagnosis and treatment capabilities at the grassroots level, there are still deficiencies in the early identification and early warning of acute exacerbation of Chronic obstructive pulmonary disease (COPD), often resulting in a high prevalence rate and high burden, but the screening rate is relatively low. In order to gradually improve this situation. In this paper, this study develop a Causal Heterogeneous Graph Representation Learning (CHGRL) method for COPD comorbidity risk prediction method that: a) constructing a heterogeneous Our dataset includes the interaction between patients and diseases; b) A cause-aware heterogeneous graph learning architecture has been constructed, combining causal inference mechanisms with heterogeneous graph learning, which can support heterogeneous graph causal learning for different types of relationships; and c) Incorporate the causal loss function in the model design, and add counterfactual reasoning learning loss and causal regularization loss on the basis of the cross-entropy classification loss. We evaluate our method and compare its performance with strong GNN baselines. Following experimental evaluation, the proposed model demonstrates high detection accuracy.
Virtual Width Networks
Nov 17, 2025



Abstract:We introduce Virtual Width Networks (VWN), a framework that delivers the benefits of wider representations without incurring the quadratic cost of increasing the hidden size. VWN decouples representational width from backbone width, expanding the embedding space while keeping backbone compute nearly constant. In our large-scale experiment, an 8-times expansion accelerates optimization by over 2 times for next-token and 3 times for next-2-token prediction. The advantage amplifies over training as both the loss gap grows and the convergence-speedup ratio increases, showing that VWN is not only token-efficient but also increasingly effective with scale. Moreover, we identify an approximately log-linear scaling relation between virtual width and loss reduction, offering an initial empirical basis and motivation for exploring virtual-width scaling as a new dimension of large-model efficiency.
WideSearch: Benchmarking Agentic Broad Info-Seeking
Aug 11, 2025Abstract:From professional research to everyday planning, many tasks are bottlenecked by wide-scale information seeking, which is more repetitive than cognitively complex. With the rapid development of Large Language Models (LLMs), automated search agents powered by LLMs offer a promising solution to liberate humans from this tedious work. However, the capability of these agents to perform such "wide-context" collection reliably and completely remains largely unevaluated due to a lack of suitable benchmarks. To bridge this gap, we introduce WideSearch, a new benchmark engineered to evaluate agent reliability on these large-scale collection tasks. The benchmark features 200 manually curated questions (100 in English, 100 in Chinese) from over 15 diverse domains, grounded in real user queries. Each task requires agents to collect large-scale atomic information, which could be verified one by one objectively, and arrange it into a well-organized output. A rigorous five-stage quality control pipeline ensures the difficulty, completeness, and verifiability of the dataset. We benchmark over 10 state-of-the-art agentic search systems, including single-agent, multi-agent frameworks, and end-to-end commercial systems. Most systems achieve overall success rates near 0\%, with the best performer reaching just 5\%. However, given sufficient time, cross-validation by multiple human testers can achieve a near 100\% success rate. These results demonstrate that present search agents have critical deficiencies in large-scale information seeking, underscoring urgent areas for future research and development in agentic search. Our dataset, evaluation pipeline, and benchmark results have been publicly released at https://widesearch-seed.github.io/
A Conjoint Graph Representation Learning Framework for Hypertension Comorbidity Risk Prediction
May 08, 2025

Abstract:The comorbidities of hypertension impose a heavy burden on patients and society. Early identification is necessary to prompt intervention, but it remains a challenging task. This study aims to address this challenge by combining joint graph learning with network analysis. Motivated by this discovery, we develop a Conjoint Graph Representation Learning (CGRL) framework that: a) constructs two networks based on disease coding, including the patient network and the disease difference network. Three comorbidity network features were generated based on the basic difference network to capture the potential relationship between comorbidities and risk diseases; b) incorporates computational structure intervention and learning feature representation, CGRL was developed to predict the risks of diabetes and coronary heart disease in patients; and c) analysis the comorbidity patterns and exploring the pathways of disease progression, the pathological pathogenesis of diabetes and coronary heart disease may be revealed. The results show that the network features extracted based on the difference network are important, and the framework we proposed provides more accurate predictions than other strong models in terms of accuracy.
Graph Clustering with Cross-View Feature Propagation
Aug 12, 2024



Abstract:Graph clustering is a fundamental and challenging learning task, which is conventionally approached by grouping similar vertices based on edge structure and feature similarity.In contrast to previous methods, in this paper, we investigate how multi-view feature propagation can influence cluster discovery in graph data.To this end, we present Graph Clustering With Cross-View Feature Propagation (GCCFP), a novel method that leverages multi-view feature propagation to enhance cluster identification in graph data.GCCFP employs a unified objective function that utilizes graph topology and multi-view vertex features to determine vertex cluster membership, regularized by a module that supports key latent feature propagation. We derive an iterative algorithm to optimize this function, prove model convergence within a finite number of iterations, and analyze its computational complexity. Our experiments on various real-world graphs demonstrate the superior clustering performance of GCCFP compared to well-established methods, manifesting its effectiveness across different scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge